Purpose:
The purpose of this lab was to perform the Chelex protocol, with hopes to replicate some of the successful results that other groups had using the Chelex method before. We then prepared 3 PCRs: a negative control, positive control, and an eDNA tube using the DNA extracted from the Chelex protocol. After extracting the DNA, we prepared the electrophoresis gel to be used in the future. We used the nano drop to determine the concentration of DNA in our sample our group extracted. The goal for every group was to have a concentration of about 10-50ng of DNA.
Procedure:
Chelex Protocol
1. Tansfer 300-500 uL of dense ciliate culture to a micro centrifuge tube
2. Label the tube and centrifuge it at 6000 x gravity for 5min
3. Discard the supernatant
4. Add 200 uL of 5% Chelex to the belle and vortex for one minute
5. Add 15 uL of proteinase K
6. Incubate for 30minutes in 56 C
7. Boil for 8min in 100C water
8. Vortex for one minute
9. Centrifuge 16,000 x gravity for 3 min to pellet the cellular debris and Chelex beads
10. Remove supernatant with DNA in solution to clean micro centrifuge tube without removing Chelex bead
11. Label the top and side of micro centrifuge tube
PCR Set-Up
1. Positive Control: 1uL of DNA template, 12.5 uL of 2X Master Mix, 1.25uL of V4 stock primers, and 10.25 uL of deionized water
2. Negative Control: 0uL of DNA template, 12.5 uL of 2X Master Mix, 1.25uL of V4 stock primers, and 11.25uL of deionized water
3. eDNA: 1uL of DNA template, 12.5uL of 2X Master Mix, 1.25uL of V4 stock primers, and 10.25uL of deionized water
40mL of 1.8% Argose Gel:
1. Pour 40mL of 1X TAE solution into a flask
2. Pour 0.72g of argues gel
3. Heat the gel until it is clear in the microwave, then allow it to cool for about five minutes
4. Pour gel into tray
5. Label tray
6. Allow the gel to cool
Nanodrop
1. Take tray containing all PCR reaction and extracted Chelex to the nano drop
2. Place 1uL drop of chDNA on node
3. Run the nano drop
4. Record concentration, 260/280 value, and the 260/230 value
5. If the concentration is not within 10-50ng of DNA, dilute eDNA reaction by adding 1uL of solution containing 5uL of water and 5uL of chDNA
Data & Observations
• Mass of Agarose gel: 1.8 x 0.4= 0.72
• PCR tubes labeled: 21-1+. 21-1 -, and 21-1 e
• chDNA labeled: TCA 21-1, chDNA with the date (4-5-18) on the side of the tube
• Nanodrop reading
◦ Concentration: 138.2ng
◦ 260/280: 0.79
◦ 260/230: 0.20
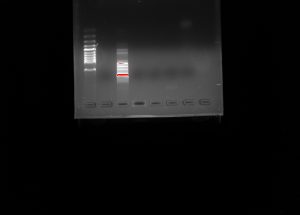
◦ Group diluted the chDNA more and results will be given next lab• Electrophoresis Gel
◦ Gel labeled 21-1
Conclusion and Future Steps
After performing the nandrop, the concentration was found much higher than the intended range. Consequentially, we had to dilute the eDNA more, however another nano drop test was not performed after our dilution due to time restrictions. In the future lab, we will run another nano drop to see if more dilutions need to be performed. We will also be running the electrophoresis through the gel. We will analyze the gels in hopes of better success than compared last time.