Lab 7: Toxicity Assay of BPA
Purpose: The purpose of part one of this lab is to make an assay of BPA using an experimental design and to determine the effects on the Tetrahymena when different concentrations of BPA are added to the stock, which will be completed by serial dilutions.
Introduction: This is the first part of the large research lab that we will be completing this semester on how UV blockers, in this case BPA, will effect Tetrahymena. Overall, we also want to look at the environment and how it will be effected by this particular UV blocker as well. If the Tetrahymena die with more BPA added to it, then that means BPA could have a negative effect on the environment, particularly with sunscreen. In this experiment, the dependent variable will be the ciliate count, the independent variable will be the UV filter, BPA, and the control will be the media that the ciliates are in. There will be three replicates per treatment, which will then be averaged; from there, we can then find the number of cells/mL in a solution by also multiplying by the dilution factor from the well that we decided to count. An assay is how you set up a design, so in this case, we will do a 1:10 dilution of 2 M BP-3. The treatment will be the UV screen BPA, as mentioned previously. BPA is 2-hydroxy-4methoxybenzophenone, with a molecular weight of 228.25 grams, where BP-3 is also known as benzophenone-3.
Materials:
- Distilled water
- Test tubes
- Wells
- Dissecting microscope
- 1 x 10^3 cells/mL solution
- Tetrahymena stock
- BP-3 (benzophenone-3)
- Glass slides
- Large pipettes
- Pipette covers
- Iodine
Procedure:
–Culture and Media Preparation:
- The Tetrahymena culture is set up and ready to be used for the experiment.
- Each group will have an aliquot of the stock culture and a tube of sterile proteose-peptone-tryptone (PTP) media and a tube of distilled water.
–Serial dilutions of the stock culture:
- Use the p1000 to pipette 450 uL of PPT into 3 wells.
- Use the p200 to pipette 50 uL of a well-mixed stock culture into the 450 uL of PPT media in well 1.
- Mix by gently shaking the plate.
- Change tips and transfer 50 uL of 1:10 dilution into the 1:1000 well.
- Mix gently by shaking the plate.
- Well 1: 1:10 –> 450 uL of media (PPT) and 50 uL of stock culture.
- Well 2: 1:100 –> 450 uL of PPT media and 50 uL of 1:10 dilution.
- Well 3: 1:1000 –> 450 uL of PPT media and 50 uL of 1:100 dilution.
–Baseline stock culture cell counts:
- Add 10 uL of iodine to each well and gently shake. Then, count the ciliates on the dissecting scope.
- Use the dissecting microscopes to observe the Tetrahymena in each well.
- Choose the dilution that will allow you to count between 10-30 cells in a 10 uL drop.
- Observe the 10 uL drop under the dissecting scope using a clean slide.
- Count at least three 10 uL drops from the optimum dilution and calculate the average number of cells/mL in the stock culture. Record your work in your notebook and on the Google Sheet.
Calculation: # of cells/10 uL x dilution factor x 1000 uL/mL= cells/mL
Data & Observations:
Experimental set-up
In the first column of wells, we have A1, B1 and C1. A1 has a 1:10 dilution, B1 has a 1:100 dilution, and C1 has a 1:1000 dilution. I took 4 samples of 10 uL of A1 and counted 46, 21, 78 and 25 cells in each of the replications. The average cell count per 10 uL was 42.5 cells/10 uL, which means that it would be 4.25 cells/uL. We then multiply this number by 10 because the dilution factor is 10, thus ending up with 42.5 cells/uL. Finally, we multiply this number by 1000 uL/mL in order to end up with 42,500 cells/mL.
The average cell count for the class was 59,045.65 cells/mL, with a standard deviation of 28,317.76. In order to find how much stock per mL we need to get the final volume, we must use C1V1=C2V2. C1 is 59,045.65, V1 serves as ‘x’, C2=1000 and V2=20 uL. The final result is 0.338725 mL, which gets multiplied by 1000 uL/mL to get 338 uL. We put 338 uL of Tetrahymena in an empty container, then fill it up to 20 mL with PPT to dilute it. Each person will make an additional 6 wells, 3 with the treatment and 3 with the control, which is distilled water. Thus, each well will end up with 1 mL.
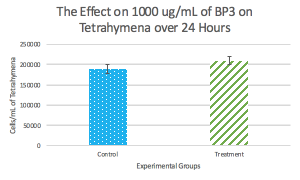
My group was assigned Treatment 4, which was 1000 ug/mL. The stock BP-3=10 mg/mL, so we must complete C1V1=C2V2 again to find the volume needed for the stock.
(1000 ug/mL)(1 mL/1000 ug)= 1 mg/mL –> (10 mg/mL)x = (1 mg/mL)(1mL), so x= 0.1 mL, or 100 uL.
These calculations show that, for our treatment, 100 uL will be the treatment, so 900 uL stock (cells) will be used to get 1000 uL of solution, which is 1 mL altogether. Thus, for column two, wells A2, B2 and C2 all contain 100 uL of the treatment and 900 uL of the stock, which is 1 x 10^3 cells/mL. In column three, wells A3, B3, and C3 all contain 100 uL of the control, which is distilled water, and 900 uL of the stock. The following day, 20 uL of iodine will be added to each of the six wells, then 10 uL drops (with three replications each) will be recorded in cells/mL.
Conclusion:
This lab allowed us to use a UV filter/blocker, BPA, to determine the effects of Tetrahymena in a stock solution and to determine what potential results could happen to the overall environment. The start time of the experiment was 2 pm (1400) and throughout the week we will check to see the results between the four different treatments and how the Tetrahymena are affected. A comparison of the treatments is crucial, because each treatment has a different concentration of BPA, which means that there should be a pattern to the amount of cells that are the solutions, depending on the particular treatment. Because our group had treatment 4, with 100 uL of treatment, we should have the least number of cells in our 10 uL observations and overall solution because we had the most treatment. On the other hand, the group that only had 25 uL of treatment should have many more ciliates in their solutions and could potentially need a serial dilution to count the cells. This lab served as the first step into the research paper that we will be writing on this entire experiment, which will continue on next week. Some potential sources of error during this lab include not shaking or mixing the solution before taking a sample and not collecting a sample from the top of the solution.