Lab 3 Isopycnic Centrifugation of Ciliates 01/24/18
Lab 3 “Isopycnic Centrifugation” 01/24/18
Pre-Lab ideas to understand:
- Ludox centrifugation
- qualities and dangers of working with Ludox SiO2
- understand process of PCR and how primers work
Purpose: Begin the process of isolating the ciliate DNA to eventually successfully barcode the genetic information.
Procedure:
- Add 8 mL of Ludox HS 40 to a 15 mL conical tube
- Quickly add 2 mL of liquid from the soil samples with a pipette into Ludox solution
- Add 2 mL of water on top of solution
- Weigh tube and label it
- Match up tube weights with another group to place into centrifuge balanced
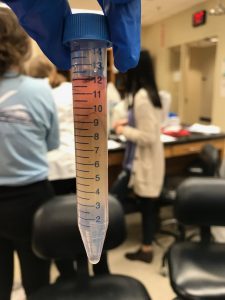
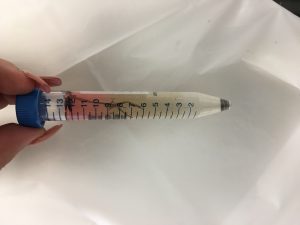
- Spin cells in centrifuge at speed 4,300 for 15 minutes
- Observe separate levels of substances in tube
- Using a plastic pipette to remove and isolate ciliate DNA and put in microfuge tube
- Label tube and place in microfuge at speed 12,000 for 1 minute
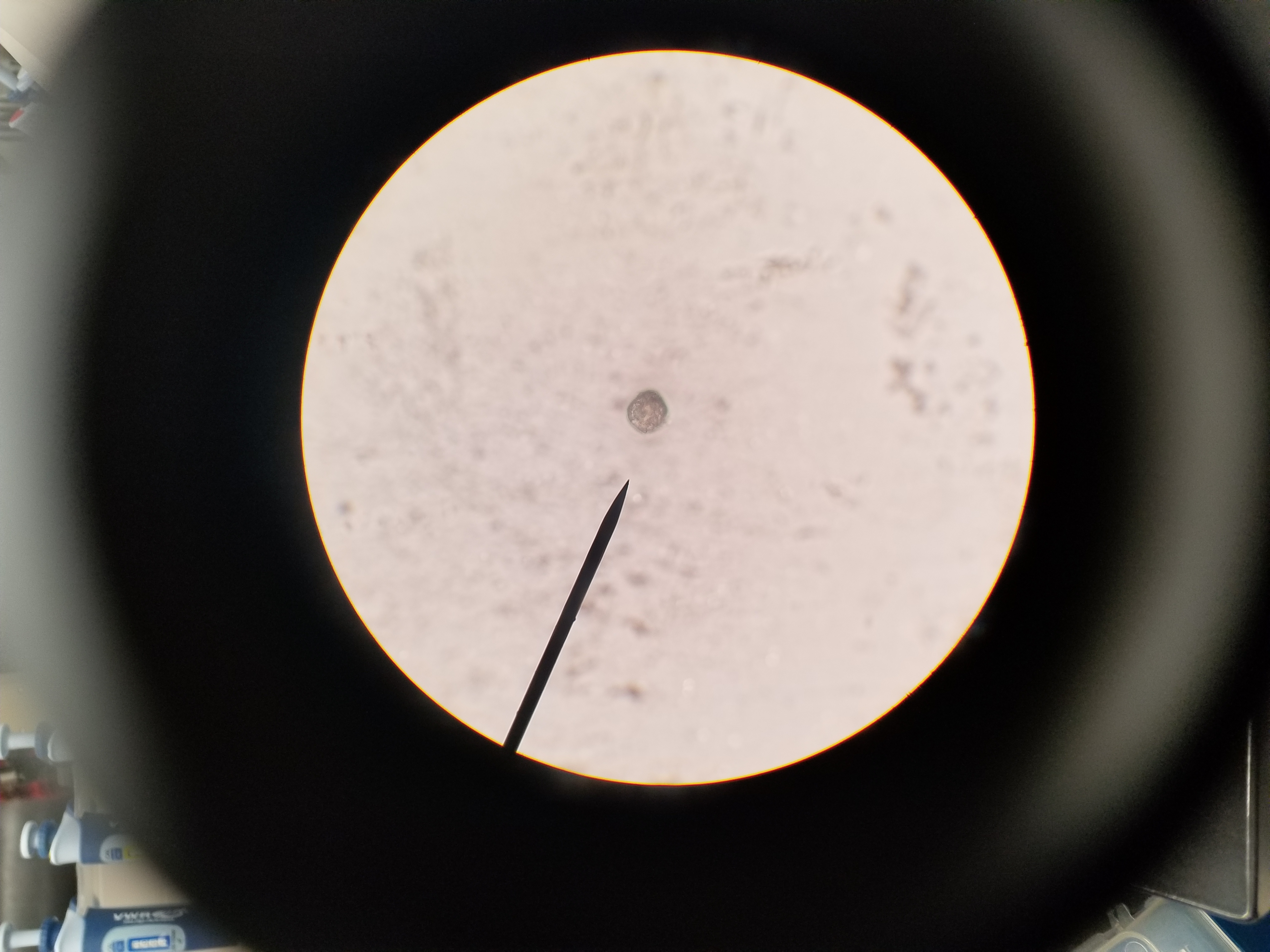
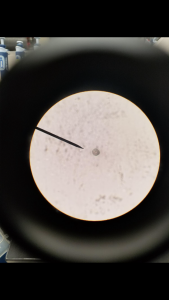
- Once DNA and dirt pellet have been separated in tube use pipette to put 2 micro liters of DNA on a concavity slide
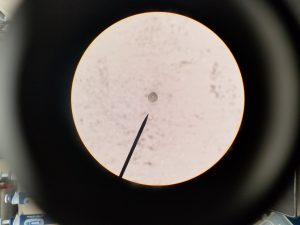
- Observe slide under compound microscope
- Freeze microfuge tube with pellet and clean off slide and return materials to appropriate location
Observations:
This is a drawing of the separated layers in our falcon tube. This is also a description of the work flow protocol we worked on throughout the lab
We had plenty of particles that were evidently DNA from ciliates that were broken up into pieces. Some looked more like membrane.
Conclusion: the Ludox solution was a successful method for separating and isolating the ciliate DNA from extraneous particles. I’m excited to see how the primers end up working and being able to look at the DNA to identify different species of ciliates. One source of error for today could have been adding the water to the top layer of the Ludox because it was difficult to control the rate at which the water came out of the pipette so it may have mixed with the Ludox more than it was supposed to. Additionally, maybe if we had sucked up more of the dirt and soil from the Bermuda grass instead of mostly water we would have more DNA particles from ciliates.
Next time: begin using the gel and primers to replicate the DNA that we captured and isolated? Still confused on how thee gel process works but I’m excited to learn about it.