1/18 research article presentations
Purpose: To explore the primary literature concerning challenges in metabarcoding soil ciliates. The purpose was also to start brainstorming protocol ideas.
Procedure (ideas for protocol):
- collect a soil sample (NOT from a flower bed/an area with a lot of artificial fertilizer)
- Dry out the soil.
- Rehydrate soil via a flooded plate.
- Isolate the ciliates in your sample (using a micropipette).
- Place the isolated ciliates in the isopycnic centriguation to separate the ciliates.
- Select a primer (rDNA?)
- Sequence the DNA using a kit (such as NextGen Sequencing)
- Amplify segments of DNA (via Amplicon)
Results (protocol):
- Compare results with other information in the data base and attempt to classify your ciliates!
Group presentations:
Group 1- The authors looked at 5 estuaries in Australia. They focused on sequencing the DNA of the protists in each estuary. An application we can use for our experiment would be the pyrosequencing because it utilizes a lot of data at one time. The Molecular Operational Taxonomic Unit could be used to cluster DNA into groups. Some challenges these authors ran into, would be the limitation of DNA based monitoring and nonproportional data.
Group 2- We can use eDNA to eliminate potentially inactive or irrelevant DNA found. Amplicon is a way for us to see similarities in the chunks of the genome in the data base.
Group 3: The protists in various places throughout Europe were sequenced in order to get a picture of the biodiversity. The challenge with this experiment was that the authors were not able to make a standard sample. Each sample was too different from the next one.
Group 4: A gene named Cox 1 was specifically singled out to depict genetic diversity between closely related protists. When a population was separated via the founder effect, the authors noticed a large variation in the Cox 1 gene. This is because the new population had its own separate gene pool. The DNA was extracted using a master purification kit which we could potentially use for our experiment.
Group 5: Several organisms (protists and ciliates) were used in this experiment. The goal was to see if the D1-D2 region was an ideal threshold for ciliates in their identification. An issue with this, is there may not be enough genetic material to work with. This article highlighted application of primers that we could use for our protocol.
Group 6: The authors of this article wanted to examine the effects of land use intensity on protists. I learned that both eDNA and rRNA are good ways to avoid obtaining unwanted inactive DNA. We can use the NextGen sequencing for our experiment as well. T-RFLP could be used after NextGen, to distinguish unique characteristics of ciliates.
Group 7: HTS (High-Throughput Sequencing) was used to sequence DNA. Dr.Adair informed us that this will not be applicable to our class because of the complexity of HTS. The issue with HTS was finding protists that matched one another.
Group 8: UniEuk is a metabarcoding system that is used to identify ciliates. The goal is to create a uniform taxonomy. Dr. Adair suggested that we start with B4 like the authors of this article.
Conclusions:
Protocols can be developed properly now that we have an idea of what to look for. We can learn from the challenges that the authors of the articles faced to make our protocol more successful. The class has developed a frame for what the protocol will look like and we have also familiarized ourselves with the terms for the equipment we will need.
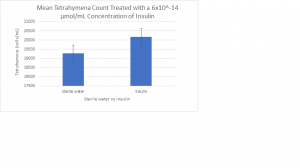
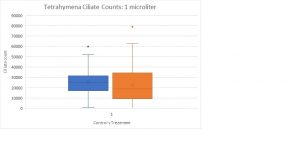
There was no sample stored.