Lab 4: Protocol Modification 2/1/18
Objective:
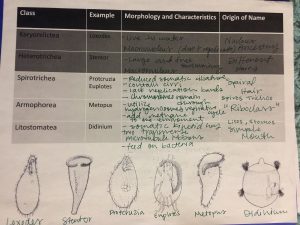
The goal of our lab is to work together as a group to incorporate all of our ideas to create a more standardized procedure that can be replicated in other research labs. By going through this process of creating a more efficient protocol, we hope to be able to successful extract DNA from ciliates and classify them.
Purpose:
The purpose of today’s lab is to come up with new ideas in order to create a more efficient Ludox protocol that would allow us to continue our ciliate metabarcoding process. We hope to create a more standardized protocol with a control that will allow us to compare the Ludox process to samples that are positive for ciliates and our samples.
Procedure:
*We came together as a class and shared our ideas for a modified Ludox protocol.
Modified Ludox Protocol-
1.) Weigh out 5 grams of soil in a glass jar and make sure you screen in for large debris.
2.) Add 7mL of water and mix for 5-10 minutes using the vortex.
3.) Pipette (cut the the tip of the pipette tip so you can get more of the sample without it getting clogged with debris) 1800 micro-liters of the watered down soil sample to mix with 200 micro-liters of a 25% glutaraldehyde solution and vortex briefly.
4.) Quickly inject 2mL of the fixed sample into the Ludox tube by placing a p1000 micro-pipette tip at the 6mL mark on the tube to ensure the soil gets into the Ludox.
5.) Carefully layer the top of the Ludox with 2mL of distilled water that has been dyed with one drop of red food coloring.
6.) Label your test tube.
7.) Centrifuge at 4300 x g for 15 minutes.
8.) Remove 2mL of the organic layer of soil from the tube using a pipette.
9.) In order to see how many cells are present, pipette out 5, 2mL drops onto a concavity slide and count all ciliates presents. Take an average of how many cells were present within the the five drops and record it to later be typed in a class data sheet.
10.) Then use the table top centrifuge and spin the 2mL tubes at 15,000 x g for one minute.
11.) Remove 1000 micro-liters of supernatant with a p1000 micropipette twice so you get a total of 2000 micro-liters. Leave a small layer of liquid in the tube to ensure you don’t pipette out the pellet (5-10 micro-liters is okay).
12.) Freeze the DNA for extraction (stored in freezer).
Note: In lab today, we only got up to step 7 and we will continue the procedure during the next lab session.
Control Procedure:
1.) Add five grams of autoclaved soil into a jar.
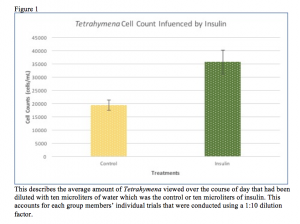
2.) Add 2mL of the Tetrahymena culture (about 2000 cells) and 5mL of water into the the jar containing the soil.
3.) Calculate the extraction efficiency: starting [cell] – extracted [cell] / starting [cell].
Data/Observations:
Test Tube Weight: 20.8 grams
Weight of Soil in Jar Sample: 5.0 grams
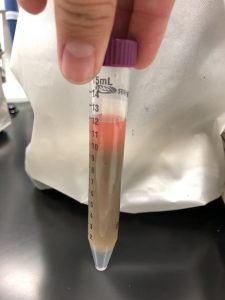
Test Tube Contained: 8mL of Ludox
I noticed that our test tube did not have as distinct separation between the ludox and the soil sample as other groups did. The soil sample and the ludox seemed to mix with each other, even after letting the sample settle, the liquids did not separate.
Storage:
Our test tube is currently labeled “G #5” to symbolize “group number five” and was placed in a yellow rack with all of the other tubes to begin centrifugation. In order to be put in the centrifuge we had to make sure that the other group that sits at the same table as ours had a test tube weight within .1 grams of ours. Once they were the same weight or within this amount then we were able to place them in this rack that was on the back lab counter closest to the computer.
Future Goals:
In the future we hope to establish a protocol that can be easily replicated and used in many other experiments related to metabarcoding. Due to the fact that today was more of an editing day and trying out part of our new procedure, we are trying to see if our new modified protocol will work better than our previous procedure. If it works we can continue the process but if it doesn’t give us the results we want we will most likely go back to the drawing board and brainstorm more ideas that we think could give us the results we want.