Lab 11: Gel Electrophoresis 3/29/18
Objective:
The objective of lab today in to run our 6 PCR samples and a DNA ladder on the 1.8% gels that we made last week. In order to successfully perform gel electrophoresis we must carefully pipette our samples into the wells without damaging them. The goal of this is so that we are able to determine whether or not the eDNA sample we are using contains the cox1 and v4 genes we were looking for.
Purpose:
The purpose of this experiment is to test our knowledge of what we have learned this far in lab. We had to calculate the amount of agarose we needed to make a 1.8% gel and we continue to do mathematical equation so that we may become more familiar with these procedures that we will have to do for the majority of our careers. We are also doing this so that we can successfully run gel electrophoresis, and if the gel is too think or heavy concentrated with agarose then it may prevent the strands of DNA from properly traveling through the gel.
Procedure:
1.) Obtain the gel created from the previous lab and the negative controls, positive controls, and eDNA samples that your group created for each primer and ladder.
2.) Place the gel into the gel box and fill the box with TAE buffer until it is just above the gel.
3.) Pipet 5 micro-liters of the primer into the first well and 10 micro-liters on the controls and eDNA samples into the following wells.
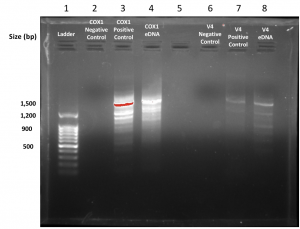
4.) More specifically, pipette the cox1 negative control into well 2, the cox1 positive control into well 3 and the cox1 eDNA sample into well 4.
5.) Then (leave well 5 empty) pipette the v4 negative control into well 6, the v4 positive control into well 7 and the v4 eDNA sample into well 8.
6.) Once all wells are full, cover the box with its lid and plug the red and black cables into the machine that conducts the electrical current.
7.) On this machine, set the voltage to 95 and start gel electrophoresis. Allow the gel to run for at least 30 minutes.
8.) Once the gel is done, remove it and place on the the BioRad Gel Doc imaging machine so that we may be able to see the bands on the gel. Record the information in your lab note book.
—————————————————————
***While waiting for the gels to complete gel electrophoresis, learn about NextGen Sequencing and the main steps of bridge amplification as well as draw a flow cell and label the oligonucleotides.
Data/Observations:
At the beginning of this lab we noticed that our original gels had been frozen so we borrowed a gel from the previous lab and Dr. Adair made them new ones. My group and I were also excited to see that both genes were found in our environmental DNA samples and we can’t wait for them to sent off and sequenced and the image above is a depiction of our gel electrophoresis results.
Storage:
The samples (previously labeled during last lab) used during gel electrophoresis were placed back in the tube rack that they were originally retrieved from since we had such great results from our gels, we were told to keep the original samples. Due to the fact that we now had an image of the gel, we were able to safely discard the gels and the gel mold was bleached and washed so that it may be reused.
Future Goals:
In the future, I hope that we able to discover new ciliates or at least be able to identify which ciliates are present within our sample if their DNA has already been sequenced. I hope to further my knowledge of the procedures we are currently using so that I may apply it to my future research. I hope that this experiment, and our results may pave the way for future labs to come. Maybe our research and the protocol we are trying to created can help scientist to further taxonomic identification of unknown ciliates.