Lab 6: E.Z.N.A Tissue DNA Kit Protocol 2/15/18
Objective:
The objective of today’s lab is to finish our Ludox protocol and start the E.Z.N.A Tissue DNA Kit protocol. We hope to be able to have successfully isolated our organic cell layer from the original sample into a centrifuged pellet that can be washed with PBS buffer to further our process of DNA extraction. We aim to continue with the procedure we had previously discussed and are prepared to adjust when something doesn’t go exactly as planned.
Purpose:
The purpose of finishing the Ludox protocol is that we may be able to start the new protocol related to PCR. The purpose of the new protocol is to perform PCR and denature the DNA so that we can run gel electrophoresis. By performing these procedures, we hope to get one step closer to being able to our goal of metabarcoding and being able to classify and analyze ciliate DNA.
Procedure:
Finishing the Ludox Protocol
1.) Centrifuge at 4300 x g for 15 minutes in a swinging bucket rotor.
2.) Remove the liquid from the cell layer, about 1.5mL below the water-ludox interface. Make sure you move a total of 4mL. Transfer this to two labeled 2mL micro-centrifuge tubes.
3.) Centrifuge the 2mL tubes at 3000 x g for 5 minutes in order to completely pellet the cells.
4.) Remove the supernatant with a p1000 micro-pipette without disturbing the pellet and dispose of the liquid in your waste container.
5.) Add 100 micro-liters of the phosphate buffer saline (PBS) to each pellet and re-suspend them by flicking and pipetting up and down. Combine the cell suspension pellets together into one tube for a total of 200 micro-liters.
6.) Place five, 2 micro-liter drops on a concavity slide and count each cell present using the 40x lens. Record your number of cells per micro-liters, then obtain a class average of cell numbers. Use iodine to see the cysts of cells under the microscope.
Beginning E.Z.N.A Tissue DNA Kit Protocol
-
Preparing Cell Suspension
1.) Wash the cells by adding 200 micro-liters of PBS buffer and re-suspend the cells with the vortex. Remove the supernatant carefully and then add 25 micro-liters of OB Protease solution, vortex briefly.
-
Lysis
2.) Add 220 micro-liters of BL buffer, pipet slowly because the solution is very viscus. Vortex briefly and incubate the tube at 70 degrees Celsius for 10 minutes in the heat block and vortex again once during this time.
-
Binding
3.) Add 220 micro-liters of 100% ethanol to the tube and vortex briefly. This will help the DNA to bind to the silica column.
4.) Insert HiBind. DNA mini column into the 2mL tube.
5.) Transfer the entire sample from step two to the HiBind. DNA mini column including any predicates that may have formed the centrifuge at maximum speed for 1 minute.
6.) Discard the liquid that went into the collection tube so you are able to reuse it.
-
Wash and Dry
7.) Add 500 micro-liters of the HBC Buffer to the column and centrifuge at maximum speed for 30 seconds.
8.) After the HBC buffer wash, discard the filtrate and collection tube and insert the HiBind. DNA mini column into a new 2mL tube.
9.) Add 700 micro-liters of DNA wash buffer the centrifuge again at maximum speed for 30 seconds.
10.) Discard the filtrate and reuse the collection tube, repeat steps 7-9 for a second DNA wash with the HBC buffer.
11.) Centrifuge the empty HiBind. DNA mini column at maximum speed for two minutes to dry the column.
-
Elute
12.) Transfer the HiBind. DNA mini column into a nuclease-free 1.5mL micro centrifuge tube and make sure the label the tube with either your name and group number.
13.) Add 100 micro-litersof elution buffer that has been heated to 70 degrees Celsius then let it sit at room temperature for about 2 minutes.
14.) Centrifuge at maximum speed for 1 minute, the discard the column and keep the 2mL tube.
15.) Store eluted DNA at -20 degrees Celsius.
*My group had to stop at step 4 because it took so long to complete the rest of the Ludox protocol.
Data/Observations:
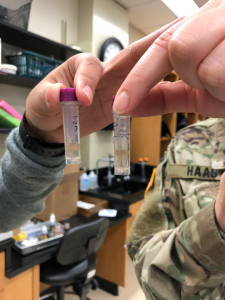
The above picture shows our 2mL tubes containing our sample and PBS buffer. My group ran into a couple problems because as you can see in our picture that the cell layer is not a pellet and looks more like debris. This photo was taking after the tube had been centrifuges twice for five minutes at 3,000 x g. The pellet would not form at the bottom and kept floating up to the top. It seems like the power of the centrifuge was not strong enough to pull the cell down to the bottom. After the cells were washed with PBS buffer about six times and centrifuged each time, Dr. Adair finally got the pellet to form at the bottom of the 2mL tube. Later, we were finally able to start washing the pellet with PBS buffer and start re-suspending the pellet and continue the procedure.
Current Storage:
The 2mL tube is labeled “LKS G# 5” and was put in a yellow test tube rack and was stored in the freezer for later use. We did not complete out experiment and stopped at step seven of the E.Z.N.A protocol.
Future Goals:
I hope that this protocol was successful and we are able to successfully extract viable DNA. Hopefully the DNA is denatured enough to perform gel electrophoresis and get results that we may be able to view. I also hope that after gel electrophoresis we can see different DNA markers and DNA of different types of ciliates. I am also excited to see how we are able to continue this procedure to reach the metabarcoding process.