Goal – Annotate genes 44, 46, 48, and 50
Tools – Phamerator, Starterator, DNA Master, GeneMark, Glimmer, NCBI BLAST, PhagesDB BLAST, HHpred
Results:
Gene 44 –
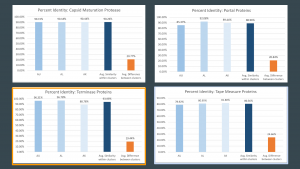
Start: 26673bp Stop: 26855bp FWD GAP: 4bp Overlap SD Score: -6.571 (2nd best score) The best score would have caused too big of a base pair overlap with the previous gene Z-Value: 1.08 CP: The gene is covered SCS: Agrees with Glimmer, Disagrees with GeneMark Genemark did not call the gene. NCBI BLAST: hypothetical protein LAROYE_46 [Arthrobacter phage Laroye]; Q30, S98 E-Value: 4e-10 CDD: No good hit PhagesDB BLAST: Laroye_46, function unknown; Q30, S98 E-Value: 1e-12 HHPred: No good hit LO: No Longest ORF would cause an overlap of 87bp ST: Agrees with Starterator F: NKF FS: NCBI, Phamerator, HHPred Notes:
Coding Potential Gene 44

NCBI BLAST 44

Gene 46 –
Start: 27430bp Stop: 27849bp FWD GAP: 299bp Gap SD Score: -4.18 (Best score) Z-Value: 2.576 CP: The gene is covered SCS: Disagrees with Glimmer, Agrees with GeneMark NCBI BLAST: No good hit CDD: No good hit PhagesDB BLAST: Waltz_Draft_46, function unknown; Q1S1 E-Value: 3e-69 HHPred: No good hit LO: Yes ST: Agrees with Starterator F: NKF FS: NCBI, Phamerator, HHPred Notes:
PhagesDB BLAST Gene 46

Gene 48 –
Start: 27837bp Stop: 28040bp FWD GAP: 13bp Overlap SD Score: -4.537 (2nd best score) The best score does not cover all the coding potential. Z-Value: 2.476 CP: The gene is covered SCS: Disagrees with Glimmer, Disagrees with GeneMark These calls left uncovered coding potential. NCBI BLAST: hypothetical protein SALGADO_37 [Arthrobacter phage Salgado] E-Value: 6e-46 CDD: No good hit PhagesDB BLAST: Edmundo_Draft_50, function unknown, Q4, S1 E-Value: 6e-26 HHPred: No good hit LO: No Longest ORF would cause an overlap of 193bp ST: Starterator was basing this call only on drafts and therefore was not very reliable. The gene was extended against Starterator to cover the coding potential F: NKF FS: NCBI, Phamerator, HHPred Notes: The start codon called by both GeneMark and Glimmer left uncovered coding potential. I chose to extend the gene to cover the coding potential.
PhagesDB BLAST Gene 48

Gene 50 –
Start: 28231bp Stop: 28398bp FWD GAP: 4bp Overlap SD Score: -5.401 (2nd best score) The best score has an ORF length of 63 bp. Z-Value: 1.624 CP: The gene is not covered There is no start codon by which to extend the gene to cover the coding potential. SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: No good hit CDD: No good hit PhagesDB BLAST: Waltz_Draft_48, function unknown, Q1 S1 E-Value: 7e-19 HHPred: No good hit LO: Yes ST: Agrees with Starterator F: NKF FS: NCBI, Phamerator, HHPred Notes:
PhagesDB BLAST Gene 50 –

Coding Potential Gene 50 –

Conclusions: Today Nihi and I finished annotating our portion of Shroom’s genome. The rest of our team has just about finished their portion too and so the next task is to review the annotations, write a cover sheet, and form a final DNA Master file for submission.