Joshua Baker
02/15/17
Today we annotated genes 4-11.
Tools used and/or Methods: DNA Master, PhagesDB BLAST, NCBI BLAST, Starterater, and HHPred.
Results:
Gene 4
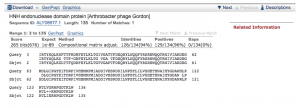
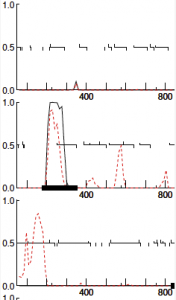
Start: 1620bp Stop: 2729bp FWD GAP: 20bp Gap SD Final Value: SD Score: -2.077 (Best score) Z-Value: 3.188 CP: The gene is not covered The gene can’t be extended to cover all – approximently 20 bp left uncovered. SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: endolysin [Arthrobacter phage Gordon] Q:1 S:1 E-Value: 0.0 CDD: PGRP and Cpl-7 domain-containing protein E-Value: 2.2e-13 PhagesDB BLAST: Gordon_4, endolysin Q:1 S:1 E-Value: 1e-164 HHPred: Endolysin, putative lys E-Value: 2.3e-23 LO: Yes ST: Agrees with Starterator F: FS: HHpred, NCBI, PhagesDB Notes: Endolysin; PGRP and Cpl-7 domain-containing protein






Gene 5 POSSIBLE DELETION

Gene 6 POSSIBLE DELETION
Gene 7
Start: 2886bp Stop: 3269bp FWD GAP: 8bp Overlap SD Final Value: SD Score: -2.904 (Best score) Z-Value: 2.767 CP: The gene is not covered The gene can’t be extended to cover all – approximently SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: hypothetical protein GORDON_5 [Arthrobacter phage Gordon] Q:6 S:39 E-Value: 2.0e-50 CDD: No good hit PhagesDB BLAST: Gordon_5, function unknown Q:6 S:39 E-Value: 6e-41 HHPred: No good hit LO: Yes ST: We cannot shorten the gene without losing the best z-score, SD value, and longest open reading frame F: NKF FS: NCBI, PhagesDB Notes:




Gene 8
Start: 3266bp Stop: 3793bp FWD GAP: 4bp Overlap SD Final Value: SD Score: -3.207 (Best score) Z-Value: 3.047 CP: The gene is not covered The gene can’t be extended to cover all – approximently SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: hypothetical protein GORDON_6 Q:18 S:1 E-Value: 2e-74 CDD: No good hit PhagesDB BLAST: Gordon_6, function unknown Q:18 S:1 E-Value: 6e-59 HHPred: No good hit LO: Yes ST: We cannot shorten the gene without losing the best z-score, SD value, and longest open reading frame F: NKF FS: NCBI, PhagesDB Notes:




Gene 9
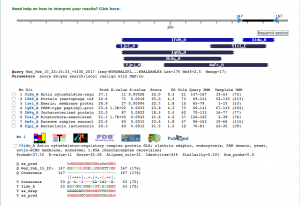
Start: 3812bp Stop: 5551bp FWD GAP: 18bp Gap SD Final Value: SD Score: -2.137 (Best score) Z-Value: 3.188 CP: The gene is covered SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: terminase large subunit [Arthrobacter phage Gordon] Q:1 S:1 E-Value: CDD: Phage terminase-like protein, large subunit, contains N-terminal HTH domain [Mobilome E-Value: 3.99e-49 PhagesDB BLAST: Gordon_7, terminase, large subunit Q:1 S:1 E-Value: 0.0 HHPred: Terminase, DNA packagin E-Value: 1.1e-27 LO: Yes ST: Agrees with Starterator F: terminase, large subunit FS: NCBI, PhagesDB, HHpred Notes:




Gene 10
Start: 5548bp Stop: 6399bp FWD GAP: 4bp Overlap SD Final Value: SD Score: -2.451 (Best score) Z-Value: 3.043 CP: The gene is covered SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: hypothetical protein CAPNMURICA_8 Q:1 S:1 E-Value: 0.0 CDD: No good hit PhagesDB BLAST: CapnMurica_8, function unknown Q:1 S:1 E-Value: 1e-152 HHPred: No good hit LO: Yes ST: Agrees with Starterator F: NKF FS: NCBI, PhagesDB Notes:





Gene 11
Start: 6396bp Stop: 6977bp FWD GAP: 4bp Overlap SD Final Value: SD Score: -4.241 (Best score) Z-Value: 2.184 CP: The gene is covered SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: hypothetical protein GORDON_9 Q:5 S:4 E-Value: 1e-79 CDD: No good hit PhagesDB BLAST: Niktson_Draft_8, function unknown Q:1 S:1 E-Value: 6e-75 HHPred: No good hit LO: Yes ST: Agrees with Starterator F: NKF FS: NCBI, Phages DB Notes:





Conclusions and Next Steps: Genes 5 and 6 were skipped over so that way we can call a group meeting and determine which one to delete. The reverse gene (5) has a better SD score and Z-value; however, the forward gene has the better start (its an ATG) and is more likely to exist being the fact that its a reverse gene. All BLAST databases don’t offer much light on the subject. Overall the other genes were easily sequenced and did not cause too much trouble. There have been a few cases were the coding potential is not entirely covered, but they are by maybe a gap of 20bp and may to the cause of some underlying genetic mutation or the result of the software. The intention is to continue annotating the genome in the next lab.