In America, “one-third of the major foodborne disease outbreaks, occurring in the U.S. between 2003 and 2007, were associated with contaminated fresh produce,” (volume 236) leaving scientists with a difficult problem; how do we combat dangerous bacterium that may be lurking on our food? Because of the fragility of most produce, harvested produce undergoes very minimal cleaning before being packaged and sent away for human consumption. An especially dangerous microorganism that can be found on produce is Escherichia coli, commonly referred to as E. Coli. A team of scientists from the University of Ohio came up with a possible solution, the use of macrophages in produce processing that will specifically target the dangerous strain of bacteria. However the question remains, is this possible and if so would the public be supportive of the idea that their produce is being washed in viral solutions?
Initially the scientists in the study decided to use E. Coli O157:H7 as the target bacterium. This bacterium can grow on the open surfaces of produce and the produce used in this study consisted of green pepper and spinach. The stain of E. Coli was first transferred to Luria Bertani Broth and incubated, spun, and incubated in a second LB broth for use in the study. Phage was isolated from cattle, sheep, and horse manure in addition to sewage from the city of Columbus Ohio. The phage was then isolated using the following procedure: samples where put into 90mL of broth and then shaken, the supernatants were then sterilized using a millipore size syringe-driven filter unit, next the samples were inoculated with the target bacterium at 35 degrees Celsius for 48 hours with shaking, lastly samples were centrifuged and filter-sterilized. The samples where then plated and individual plaques were plated a second time to isolate a single phage.
Each type of phage was tested to see which one was the most effective and the most effective phage was the one that caused the most inactivation of the target bacterium when plated in LB agar. Before the individual produce could be tested green pepper was put under an UV light for one hour on each side to kill any background microorganisms. Spinach was not put under the UV light because it was too fragile to withstand the UV treatment.
Actual testing involved dipping green pepper in crude lysate for five minutes and shaking. Spinach was also dipped in crude lysate for 2 minutes and shaken because it was too fragile to withstand the total shaking time. A control was also only dipped in E. Coli. The samples were then left to dry for a period of 4 hours at room temperature and then refrigerated for a period of 72 hours to simulate produce manufacturing. Pepper samples and spinach samples were then plated and calculations preformed. Because spinach could not be treated with UV light the bacterium used was altered to produce a green fluorescent light and the colonies were then counted.
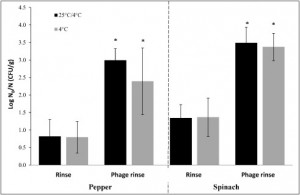
Compared to just the rinse the use of phages to combat E. Coli O157:H7 was extremely successful showing major inhibition of bacterial growth.
Overall the experiment was a success. The use of phages to combat bacterium is not only possible, but extremely effective – reducing E. Coli populations by 2.0 Log. Thus the applications of phages in produce manufacturing may soon become a reality. One problem still remains, this was small scale, further research would have to be preformed industrially and currently the scientists from Ohio are working to create that opportunity. Lastly public education would be required before this treatment can be put into practice as the majority of the American and world population for that matter knows very little about phages and the benefits that they can also have. In conclusion the experiment was a success and in the coming years we may begin to see phages integrated into our sanitation systems worldwide.
The link to, “Developing and optimizing bacteriophage treatment to control enterohemorrhagic Escherichia Coli on fresh produce,” can be found here. The article, written by Abigail B. Snyder, Jennifer J. Perry and Ahmed E. Yousef from the University of Ohio, was found using the Scopus search engine. Key words and limits used to find the article include: bacteriophage, article (limit to), 2016 (limit to) and yielded a total of 587 articles.