Meredith Kim
April 11, 2017
Rationale of today’s work: Delegate jobs to each group member and begin researching promoter sequences for the phages containing genes in pham 7442. This pham has 16 members, all of which are in the AO cluster. We will give each member about 5 genes to thoroughly investigate through DNA Master’s Promoter prediction and NCBI blast. This time, however, we will be focusing on running the promoter prediction for the gap before the gene itself. Previously, we ran the promoter prediction on the gene itself, which was not very useful since promoters are typically found previous to the actual gene.
Tools used and/or Methods: PhagesDB glossary, Phamerator, DNA Master, NCBI
Results:
Title for our Project: Life if PHAM-tastic !!
Phage Distribution:
Meredith:
- Barrett Lemon Gene #7
- Boss Lady Gene #7
- Beans Gene #6
- Brent Gene #6
- JKerns Gene #7
- Jordan Gene #7
Taylor:
- Fanzy Gene #6
- Jawnski Gene #6
- Nahla Gene #6
- Piccoletto Gene #6
- LeeroyJ Gene #7
- Timinator Gene #7
Katie:
- Martha Gene #7
- Shade Gene #7
- Sonny Gene #7
- StevieBAY Gene #7
- TaeYoung Gene #7
- Zartrosa Gene #7
For Each Gene:
- NCBI Blast Previous Gene
- Note gap between the gene and wether previous gene is forward/reverse
- Insert a gene into DNA Master in the gap between the previous gene and the gene in the pham we are investigating
- Run DNA Promoter on the gap sequence
- Copy/Paste the actual sequence of the gap containing the promoter
- NCBI Blast the Specified gene in the pham
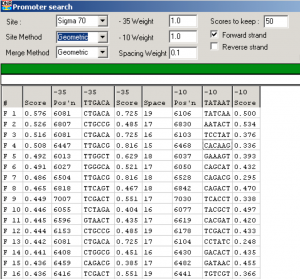
Barret lemon Gene #7:
Previous gene (#6) is a reverse gene… Gap: 128bp
Blast NCBI for Gene #6 is hypothetical protein BARRETLEMON_6 e-value: 1e-48, q:1 s:1, no CDD

|
|
-35 |
-35 |
-35 |
|
-10 |
-10 |
-10 |
| # |
Score |
Pos’n |
TTGACA |
Score |
Space |
Pos’n |
TATAAT |
Score |
| F 238 |
0.678 |
5927 |
CTGAGC |
0.592 |
17 |
5950 |
TTGAAT |
0.732 |
Gap Sequence:
TTGTCTTCTCTTCTCTCTGGCGGGTTGTTCGAACTGCTTGATCAAGTATGGACAGGCTGGCTCGTAAAGCGCAAGCCGAAACACGCGCACGTCCCACAACTGAGCCGCCCGGCACAGCACAGTTGAATCA
NCBI Blast for Gene #7: scaffolding protein [Arthrobacter phage BarretLemon], e-value: 0.0, q:1 s:1, Mu-Like_Pro superfamily
Boss Lady Gene #7:
Previous gene (#6) is a reverse gene… Gap: 134bp
Blast NCBI for Gene #6: hypothetical protein MARTHA_6 [Arthrobacter phage Martha], e-value 1e-47, q:1 S:1

|
|
-35 |
-35 |
-35 |
|
-10 |
-10 |
-10 |
| # |
Score |
Pos’n |
TTGACA |
Score |
Space |
Pos’n |
TATAAT |
Score |
| F 436 |
0.657 |
5817 |
TTCTCT |
0.633 |
15 |
5838 |
TAGAAC |
0.72 |
Gap Sequence:
TTGTCTTCTCTTCTCTCGTTGTTCGAACTGTTAGAACAAGTATGGACAGATAGATCGAGATCGCGCAACCACCCAGCAGGCTTGTTCGATCGTGTCATCCCACAACGCAGCCGCGCGGCACGTCAGAGTTGACTCA
NCBI Blast for Gene #7: scaffolding protein [Arthrobacter phage Sonny], e-value: 0.0, q:1 s:1, Mu-Like_Pro superfamily
Beans Gene #6:
Previous gene (#5) is a forward gene… Gap: 156bp
NCBI Blast for gene #5: hypothetical protein SEA_BRENT_5 [Arthrobacter phage Brent], e-value: 1e-53, q:1 s:1, no CDD

|
|
-35 |
-35 |
-35 |
|
-10 |
-10 |
-10 |
| # |
Score |
Pos’n |
TTGACA |
Score |
Space |
Pos’n |
TATAAT |
Score |
| F 166 |
0.682 |
5563 |
CTGATC |
0.614 |
17 |
5586 |
CACAAT |
0.719 |
Gap Sequence:
ATTCTCGTGTTAATGTGAGCTTGCCTGTTTGGGACACGGCTTGACTGGGGAGTCTGGGAAGGGGCGGCGTCATTGCGACGCCGCCCTTTCTTGTGCCAGCTGATCGAACACGCCCGGCGTCCCACAATCCCGCAGCTGAGCGTTTCACAGTGGAGCTA
NCBI Blast for Gene #6: scaffolding protein [Arthrobacter phage Jawnski], e-value:0.0, q:1 s:1, Mu-Like_Pro superfamily
Brent Gene #6:
Previous gene (#5) is a forward gene… Gap: 157bp
NCBI Blast for Gene #5: hypothetical protein SEA_BRENT_5 [Arthrobacter phage Brent], e-vlaue: 7e-60, q:1 s:1, no CDD

|
|
-35 |
-35 |
-35 |
|
-10 |
-10 |
-10 |
| # |
Score |
Pos’n |
TTGACA |
Score |
Space |
Pos’n |
TATAAT |
Score |
| F 182 |
0.677 |
5478 |
TTGCCT |
0.77 |
15 |
5499 |
TTGACT |
0.625 |
Gap Sequence:
ATTCTCGTGTTAATGTGAGCTTGCCTGTTTGGGAAGACGGCTTGACTGGGGAGTCTGGGAAAGGGCGGCGTCATTGCGACGCCGCCCTTTCTTGTGCCAGCTGATCGAACACGCCCGGCGTCCCACACTCGCGCAGCCGGGCGCTTCACAGTGGAGTCA
NCBI Blast for gene #6: scaffolding protein [Arthrobacter phage Brent], e-value: 0.0, q:1 s:1, Mu-Like_Pro superfamily
JKerns Gene #7:
Previous gene (#6) is a reverse gene… Gap: 134bp
Blast NCBI for Gene #6: hypothetical protein SONNY_6 [Arthrobacter phage Sonny], e-value: 3e-50, q:1 s:1, no CDD

|
|
-35 |
-35 |
-35 |
|
-10 |
-10 |
-10 |
| # |
Score |
Pos’n |
TTGACA |
Score |
Space |
Pos’n |
TATAAT |
Score |
| F 447 |
0.657 |
5814 |
TTCTCT |
0.633 |
15 |
5835 |
TAGAAC |
0.72 |
Gap Sequence:
TTTTCTTCTCTTCTCTCGTTGTTCGAACTGTTAGAACAAGTATGGACAGATAGATCGAGATCGCGCAACCACCCAGCAGGCTTGTTCGATCGTGTCATCCCACAACGGGGCCGCGCGGCACGTCAGAGTTGACTCA
NCBI Blast for gene #7: scaffolding protein [Arthrobacter phage Sonny], e-value: 0.0, q:1 s:1, Mu-Like_Pro superfamily
Jordan Gene #7:
Previous gene (#6) is a reverse gene… Gap: 134bp
Blast NCBI for Gene #6: hypothetical protein SONNY_6 [Arthrobacter phage Sonny], e-value: 3e-50, q:1 s:1, no CDD

|
|
-35 |
-35 |
-35 |
|
-10 |
-10 |
-10 |
| # |
Score |
Pos’n |
TTGACA |
Score |
Space |
Pos’n |
TATAAT |
Score |
| F 453 |
0.657 |
5814 |
TTCTCT |
0.633 |
15 |
5835 |
TAGAAC |
0.72 |
Gap Sequence:
TTTTCTTCTCTTCTCTCGTTGTTCGAACTGTTAGAACAAGTATGGACAGATAGATCGAGATCGCGCAACCACCCAGCAGGCTTGTTCGATCGTGTCATCCCACAACGGGGCCGCGCGGCACGTCAGAGTTGACTCA
NCBI Blast for gene #7: scaffolding protein [Arthrobacter phage Sonny], e-value: 0.0, q:1 s:1, Mu-Like_Pro superfamily
Conclusion: All the genes in the pham are scaffolding proteins and are preceded by hypothetical proteins. However, there was not a very clear correlation in the promoter sequences that I annotated. The other two members in my team have annotated the other 12 genes, so I need to look at those, too, when making generalizations and looking for a correlation. Our goal is to have all 18 phages in the pham blasted and their promoter sequences predicted.
Next Steps: Next week, we want to compare our results from the blasts and promotor predictions. If time allows, we will begin running various programs to determine the percent CG in the promotor sequences.