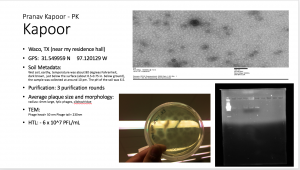
Final Lab Notbook Check
Day 5: Meeting and Collaboration
2/20/17
We held another meeting as a Nubia group as planned to discuss how far everybody was on their parts of the annotation process. After the meeting, we found out that Lathan and Dr. Adair wanted us to include the Q and S values for both PHages DB and NCBI Blast, so we re-blasted the genes we worked on. After, we finished looking at the Q an S values we checked over each others work as we were almost done.
After this, we exchanged data with Navya and Christina as they were also done with their annotations. In the time left, I checked genes 32 and 34. Gene 32 was good, but their gene 34 had a different NCBI blast and conserved domain hit from what I had. I consulted with them and they realized that they forgot to change the blast results so I went ahead and corrected that.
Day 6: More Double Checking
2/22/27
I continued checking Navya and Christina’s annotations. I checked genes 38, 40, 42, and 44. My phamerator froze my computer for a while which is why I only checked 4 genes through the lab time. Out of these 4, I discovered a problem in the HHPred e-value. I consulted with Navya and Christina and we changed the value from 7.7e^-24 to 8.2e^-24. We spent the entire day checking and double checking each other’s genes.
Day 7: Meeting
2/27/17
Today we had another team meeting. We were told to split up the entire Nubia group into separate groups to do different jobs. Alex and I were tasked with creating a Master DNA file. This DNA Master file was to include the final annotations in the description of the DNA Master file which is supposed to be a part of the final submission. We worked on this the entire lab period. I added the full and complete annotations to to the odd numbered genes while Alex added them to the even numbered genes.
Day 8: AN Phage
3/1/17
Today, Dr. Adair announced that there was AN phages that we could potentially take up. I decided to take one named Saphira. Alex volunteered to take over the full burden of the final DNA Master file which I really appreciated. I started working on Saphira. I am working on this phage with Chrissy. I have genes 1-13. I had trouble figuring out gene one as I didn’t remember what to do with a gene that on DNAMaster continued on from the end to the beginning, but I consulted with Lathan and he refreshed me on the procedure. Otherwise, I worked genes 2,3, and 4 with no further complications.
Day 9: Continuation of the annotation of the AN Phage Saphira
3/13/17
I continued annotating the phage Saphira, and completed genes 5,6,7, and 8. I also helped Alex out with checking starterator and phamerator as the program was not working on her computer.
Day 10: Meeting for group project
3/15/17
Today we had a meeting for the group project. Our Group consists of me, Micheal, Alec, and Daniel. We plan to meet this Saturday after doing some homework. We each have to look over some posters, write up a possible conclusion and introduction, and search for ideas we could use in our presentation and for our topic.
Below is a picture of what we brainstormed on Saturday(3/18/17):
Day 11-Day 12: Group Project
3/20-3/22
Our group project is due on the 22nd of March so for these lab days we used these days and out of labs to prepare for the presentation and the poster. Below is a picture of our poster:
Day 13: Scholars Week Practice
3/27/17
Today we got together in our Scholars Week Presentation. Our group is me, Micheal, Alec, and Daniel. We got assigned the poster we had to present and we worked on being able to explain it properly, especially the 16/17 gene slippage and the Gepard plot that were present on the poster.
Day 14: Presenting at Scholars Day
3/29/17
Today we spent the lab time taking shifts presenting our poster at Scholars Day in the Atrium of the BSB. It was a really cool experience I thought it was really cool that people were coming to us to provide them knowledge in the subject and that we were the experts and I wasn’t on the other side of things for once. There will always be more questions, but it felt good to educate people about our poster.
Day 15: Preparing for LaTourneau
4/3/17
Today in lab, the people who were going to attend the Regional SEA-PHAGES Symposium at LaTourneau split off from the rest of the group to discuss what the gameplan was. We got split into different presentation groups again for LaTourneau.
Day 16: Preparing for LaTourneau Continued
4/5/17
Today we presented our poster to the people in the class in order to prepare to present at the Regional SEA-PHGAES Symposium. Below is a picture of our group at LaTourneau in fromt of the poster we presented.
Day 17-Now: Individual Project
4/10/17-4/26/7
After the SEA-Phages Regional Symposium, which I thought was really fun and a great learning experience, we started working on our individual projects. Below is a sheet in my notebook that we used to plan out the project.
Thomas was very interested in exploring the potential for there to be a connection between start codons and if they code for a known function or not. So going ahead with this topic, we struggled to find a paper at first that showed that our topic was possible or feasible as there hasn’t been much research on codons. Unfortunately, a program we had hoped to use to analyze these codons didn’t work and we spent two labs trying to fix the program and make it run but it didn’t. This really set us back as we had to do a bunch of number crunching by hand and by looking at the codons ourselves for each phage in each cluster so we were not able to take the project in the direction that we wanted to.