4/19/17 Independent Project Planning Jake Hanna
Goal/Rationale:
(I was gone on the 12th and couldn’t make any contribution to the research). Noah and I will use a single gene sequencing tool to analyze the overall usefulness of three genes we found to be present shrooms, caterpillar, and nubia, as well as the TMP (tape-measure-protein) gene., which were the: Capsid Maturation Protease, Terminase large subunit, and a Portal Protein genes. Through the Single gene sequencing tool, we will be able to evaluate the conservation of genes within the clusters. Through this program we will be able to quantify Andreas Gepard-dotplot data, and get a clearer picture of how conserved the selected genes are to each other.
Tools/Procedure:
We will be using the phagesDB database to find the appropriate phages to put through the Single Gene sequencing tool, Cluster Omega, to analyze how similar the same genes are across phages of different clusters as well as within the same cluster. High percentages for similarity from the genes within indicate a low degree of conservation. A high percentage of similarities between genes that come phages of different clusters would indicate that it would be a less effective tool for single gene analysis.
Results:
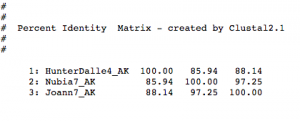
AK cluster
Findings/Next Steps:
While only going through a small portion of what we will need to analyze, these results have already begun to show a high level of inter-cluster gene conservation, but more data will have to be put together to analyze the true significance of our findings.