SEA Phage Lab Day 30
Day 30: 11/30/16 TEM
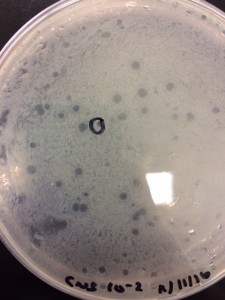
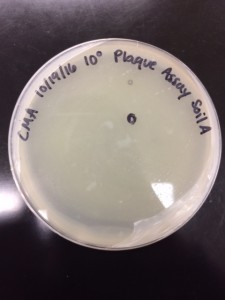
Purpose: To see my phage.
Materials: high titer lysate, parafilm, sterile water, tweezers, copper grids, 2% uranyl acetate
Procedure:
- I transferred 100 microliters of my lysate into a micro-centrifuge tube.
- I stretched out a piece of parafilm.
- Then I put 20 microliters of the lysate on the left corner of the parafilm.
- Then next to it I put 20 microliters of sterile water.
- Next to the water, I put another 10 microliters of sterile water.
- Lastly, I put 20 microliters of uranyl acetate on the right corner of the parafilm.
- After I set up my work area, I put a small circular copper sheet in the lysate for about 5 minutes so that the phages would attach to the copper sheet.
- After 5 minutes, I transferred the copper sheet to the first drop of water for 1 minute.
- After 1 minutes, I transferred the copper sheet to the second drop of water for 1 minute.
- After 1 minute, I transferred the copper sheet to the uranyl acetate for 30 seconds.
- Then I dabbed the copper sheet on a piece of paper to get rid of the excess liquid.
- Then Jennifer put my copper sheet into the section of the small rectangular box labelled 1E.
- Afterwards, I went to the TEM machine to look for my phage.
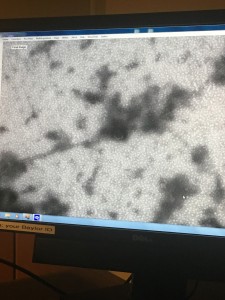
- The Dr. Burnd who was looking for the phage said that he could not find anything on my copper sheet except one that looks very weird. So he found two phages on Andrea’s plate and took pictures for me since I adopted her phage.
Conclusion: We can’t really tell in this picture where the tail ends and where the head attaches. It looks like there are two tails coming out from the head, which is not an accurate image of a phage. So, we decided that we can’t get a clear image of my phage. Although I adopted Andrea’s phage, I cannot just use Andrea’s pictures because on her plates she had plaques of different sizes so our plaque may not be the same.
Next Step: Run gel.