Date of Lab: 02/28/17
Rationale: The purpose of the lab was to annotate 5 genes from Phage Timinator.
Tools used: DNAMaster, Phamerator, Starterator, GeneMark, NCBI Blast, CDD, HHPred
Procedures: Annotate Gene 21,43,65,73(B),12(B) through the tools listed above and record the results on spreadsheet.
Conclusion:
I only worked on gene 21,43 and part of 65 in this lab. I did not make any changes on gene21,43 and 65, they were all auto-annotated.
Gene 21: According to NCBI blast and Phamerator, it is a tail protein [Arthrobacter phage BarretLemon], so it has a function of major tail protein.
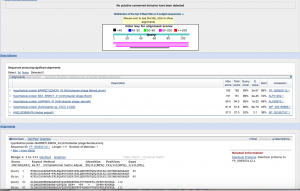
Gene 43 has no known function and has Q1S1.
Gene 65 has no known function and has Q1S1.
| 21 |
Start: 17046bp Stop: 18386bp FWD GAP: 0bp Overlap SD Final Value: SD Score: -4.943 (4th best score) longest ORF Z-Value: 2.005 CP: The gene is not covered not able to move any further and it agrees with starterator SCS: Agrees with Glimmer, Disagrees with GeneMark not able to move the starting site or that will create a huge overlap and it agrees with starterator NCBI BLAST: tail protein [Arthrobacter phage BarretLemon] E-Value: 0.0 CDD: CHAP domain; This domain corresponds to an amidase function. Many of these proteins are … E-Value: 5.83e-10 PhagesDB BLAST: No good hit HHPred: ORF30/ORF32, LYSK; vira E-Value: 1.4E-22 LO: Yes ST: Agrees with Starterator F: major tail protein FS: tail protein (NCBI), CHAP domain-amidase (phamerator) Notes: |
| 43 |
Start: 34542bp Stop: 34874bp FWD GAP: 47bp Gap SD Final Value: SD Score: -3.603 (Best score) Z-Value: 2.642 CP: The gene is covered SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: hypothetical protein BARRETLEMON_43 [Arthrobacter phage BarretLemon] E-Value: 1e-57 CDD: No good hit PhagesDB BLAST: Timinator_Draft_43, function unknown, 110 Q:1 S:1 E-Value: 2e-56 HHPred: No good hit LO: Yes ST: Agrees with Starterator F: NKF FS: NCBI, Phages DB Notes: |
| 65 |
Start: 43017bp Stop: 43394bp FWD GAP: 4bp Overlap SD Final Value: SD Score: -2.238 (Best score) Z-Value: 3.351 CP: The gene is covered SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: hypothetical protein BARRETLEMON_64 [Arthrobacter phage BarretLemon] E-Value: 5e-86 CDD: No good hit PhagesDB BLAST: Timinator_Draft_65, function unknown, 125 E-Value: 7e-70 HHPred: No good hit LO: No it will create a large overlap if we choose the one with longest ORF ST: Agrees with Starterator F: NKF FS: Phages DB, NCBI Notes: |
If there is no other starting site close to the auto annotated starting site and the gene was not fully covered(on geneMark), just keep the starting site unchanged even though it was not fully covered. If not it will create a huge overlap with the gene in front.
gene43

gene21

gene65 NCBI blast

gene65 HHpred















