Lab 15: Poster Presentations
Taylor Hutcheson
Date:
April 28, 2017
Abstract
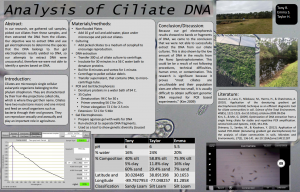
In our research, we gathered soil samples, picked out ciliates from these samples, and then extracted the DNA from the ciliates. Our objective was to extract DNA and use gel electrophoresis to determine the species that the DNA belongs to. Our gel electrophoresis results yielded no DNA, so our attempts to extract DNA were unsuccessful, therefore we were not able to identify a species based on DNA.
Introduction
Ciliates are microscopic single cellular eukaryotic organisms belonging to the phylum ciliophoran. They are characterized by their hair-like projections called cilia, which is where they get their name. Ciliates have two nuclei (one macro and one micro) and feed on small organisms such as bacteria through their oral grooves. They can reproduce sexually and asexually and play an important role in agriculture.
Materials/Methods
- Non-flooded Plates
- Add 10 g of soil and add water, place under microscope and pick out ciliates.
- Culturing
- Add picked ciliates to a medium of ceraphyll to encourage reproduction.
- DNA extraction
- Transfer 300 uL of ciliate culture to centrifuge.
- Incubate for 30 minutes in a 56 C water bath to denature proteins.
- Boil for 8 minutes and vortex for 1 minute.
- Centrifuge to pellet cellular debris
- Transfer supernatant, that contains DNA, to microcentrifuge tube.
- PCR and Gel Electrophoresis
- Denature proteins in a water bath of 94 C.
- 35 Cycles
- Denaturation: 94 C for 30 s
- Primer annealing 56 C for 20 s
- Primer elongation 72 C for 2.5 min
- Extension: 72 C for 5 Min
- Gel Electrophoresis =
- Prepare agarose gel with wells for DNA
- Run electrical to separate DNA fragments.
- Used as a tool to show genetic diversity (Jousset 2010)
Conclusion/Discussion
Because our gel electrophoresis results showed no bands or fragments of DNA, we came to the conclusion that we were not able to successfully extract the DNA from our ciliate cultures. This is also shown by the low amount of DNA in the results from the Nano Spectrophotometer. This could be be a result of not following procedures, technical difficulties, human error, or contamination. This research is significant because it proves “most ciliates are uncultivable and their population sizes are often too small, it is usually difficult to obtain sufficient genomic DNA required for PCR based experiments” (Kim 2009).
Literature
Jousset, A., Lara, E., Nikolausz, M., Harms, H., & Chatzinotas, A. (2010). Application of the denaturing gradient gel electrophoresis (DGGE) technique as an efficient diagnostic tool for ciliate communities in soil. Science of the Total Environment, 408(5), 1221-1225.
Kim, S., & Min, G. (2009). Optimization of DNA extraction from a single living ciliate for stable and repetitive PCR amplification. Animal Cells and Systems, 13(3), 351-356.
Shimano, S., Sambe, M., & Kasahara, Y. (2012). Application of nested PCR-DGGE (denaturing gradient gel electrophoresis) for the analysis of ciliate communities in soils. Microbes and Environments, 27(2), 136-141.