FINAL NOTEBOOK CHECK
Date: May 1, 2017
Classifying Ciliates using DNA Extraction
FIGURE 1:
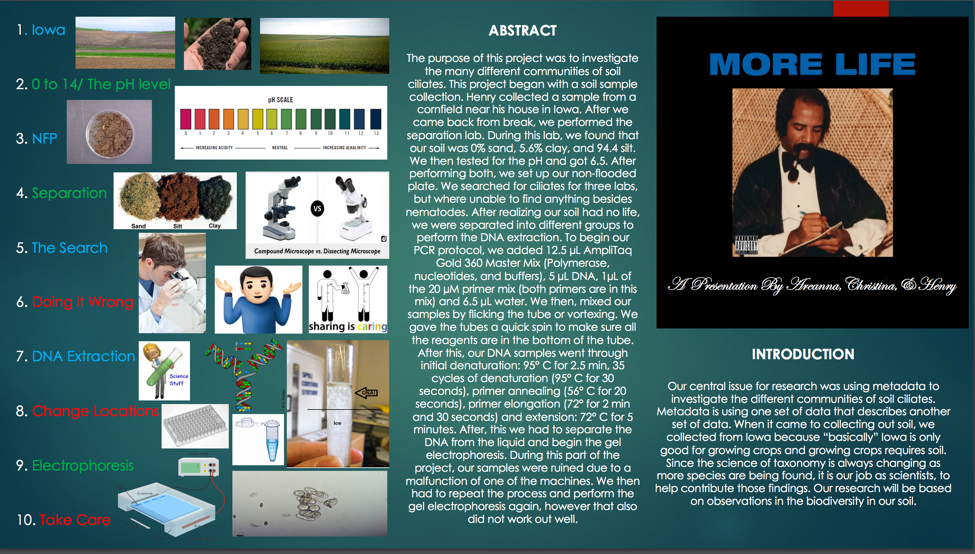
In Figure 1 we have our final poster project. This summarizes what we have been doing over the past few weeks, while classifying ciliates.
Procedures:
Soil and Metadata collection:
- Collected a soil sample from Iowa in a corn field because Iowa has rich soil.
- Record information about our soil such as, location (GPS), date, time of day, temperature, last rainfall, humidity, color, etc..
- Location/Climate
- Iowan Cornfield
- Latitude: 41.470282
- Longitude: -95.318582
- Date: March 11, 2017
- Time: 6:30 pm
- Temperature: 29 degrees F
- Humidity: 65%
- Last Rainfall: 1 month prior
- Color: Brown
- Location/Climate
- Then transferred 5 g of the sample into a weighing boat, and weighed the new sample.
- Wait 1 week and weigh the boat again.
- Calculate the percent composition of the sand, silt, and clay.
- % Sand –> 0%
- % Clay –> 5.41%
- % Silt –> 94.59%
- Obtain the pH of the dirt with pH paper
- Ours was pH was 6.5 = acidic
- Added soil to 5mL mark of the falcon tube and added water until the 10mL mark. Then I vortexed the mixture.
- This is to help find what type of soil it is.
- After a week, the falcon tube divided the mixture into layers.
- Measure the layers using a ruler and do some calculations to get a percent composition.
- Our dirt was mainly silt.
- Next I took the soil sample and placed it into a non-flooded dish, and put just enough water to where there is some runoff, but didn’t flooding it. Then observed under a microscope.
- After three days of searching, I found no sign of ciliate life.
DNA Extraction:
- I borrowed my new sample from the infamous Lily because she knew how to raise her ciliates.
- Her % composition was:
- Sand–>92%
- Silt–>0.4%
- Clay–>8%
- Her % composition was:
- Transfered 400uL of her cultured ciliates into a centrifuge tube.
- Centrifuged the tube for 5 minutes at 6000g.
- I Added 200uL of Chelex into the tube and vortexed for 1 minute
- Incubate at 56 degrees C for 8 minutes
- Then place the tube in a 100 degrees C water bath for 8 minutes.
- Centrifuge at 16000g for 3 minutes.
- Transfer the supernatant into another centrifuge tube and label with the Soil identifier
- HHN0617
- Then to determine the purity of the sample, a nanodrop spectrometer was used.
PCR:
- 12.5 uL 2x Master Mis, 5uL DNA, 1uL of uM primer and water was added to form a PCR mixture.
- It was added to the 25uL volume mark.
- The control tube didn’t have DNA but had more water, while the experimental tube had the more DNA
- Label the control tube 6OC and the experimental tube 6O.
- Both of the tubes were vortexed for 10 seconds and stored until the lab was over.
Gel Electrophoresis:
- We added 100 ml of 1XTAE and 1.5 g of argose to a 250 mL flask and microwaved.
- We assembled the tray by placing the comb into the tray with the two bumpers on each side.
- We filled the tray with gel about halfway up the combs.
- Thermocycling procedure:
- Denaturation at 95 degrees C for 30s
- 35 cycles:
- Denaturation at 95 degrees C for 30s
- Annealing at 56 degrees for 20 s
- Elongation at 72 degrees C for 2:30s
- Extension at 72 degrees C for 5 minutes
- Added 5 ul of the ladder to the product using a micropipet into the gel tray.
- We then placed the lid on the tray after loading the DNA samples into their respected loading zones, and turned it on at 100 volts for 30 minutes
- We placed the tray under UV light so we could actually see the results.
Here is a visual representation of our Gel Electrophoresis:
Labeling Info:
- Soil Identifier: HHN0617
- PCR Label: 6O
Ideas for Future Experiments:
- Make sure to double check your work, even if you think you did it right the first time.
- Don’t be afraid to question the results or the process
- Use more than one sample



