Phage Annotation #3 09_12_17
Background: We have currently been working through the process of annotation and understanding the different tools/programs involved in our annotation process. We have annotated Link Gene 2 in class in order to reinforce this process.
Purpose: Master the ability to annotate the structural features of a predicted gene. Master the knowledge of the meanings for each shortcut in the annotation template. Explore the use of Phamerator to discover whether a gene function is conserved and compare start sites of published genomes. Explore the mosaic nature of phage genomes using Phamerator and analyze the differences between closely related genomes. Use BLASTp and the CDD to explore preciously predicted gene function.
Procedure:
- Annotation Review and Practice (save in notebook/blog for later use)
- Open your saved auto-annotated Link DNAMaster file
- Check your DNAMaster settings
- Open the Guiding Principles and use to assist in annotation (remember to use the template SSC: GAP: CP: SD: Z: SCS: NCBI Blast: PDB BLAST: HHPred: LO: ST: F: FS:)
- Use GeneMark for CP, NCBI for NCNI Blast, and phagesdb for PDB
- Annotate Gene 9 on your own
- Find the most likely start codon instead of the default start codon setting
- Introduction to Phamerator
- Practice using Phamerator with annotated gene
- Introduction to CDD (Conserved Domain Database)
- Annotation practice
Results:
Conclusion:
In lab we have fine tuned our annotation process for most of the template that we are using.
- Link Gene 3
- SSC: Start: 591 Stop: 806 GAP: 30 CP: Covered SD: -6.444 Z: 1.190 SCS: No, we chose the longest ORF NCBI Blast: hypothetical protein hypothetical protein SEA_DECURRO_3 [Arthrobacter phage Decurro] E-value: 2e-46 PDB BLAST: TymAbreu_3, function unknown E-value: 9e-41 LO: Yes
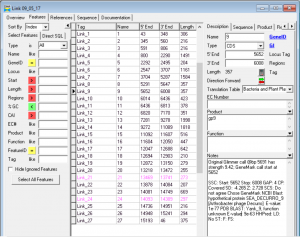
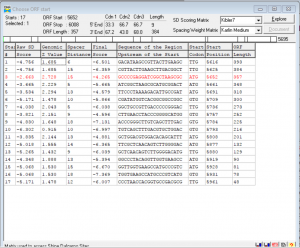
- Link Gene 9
- SSC: Start: 5652 Stop: 6008 GAP: 4 CP: Covered SD: -4.265 Z: 2.728 SCS: Do not agree Chose GeneMark NCBI Blast: hypothetical protein SEA_DECURRO_9 [Arthrobacter phage Decurro] E-value: 1e-77 PDB BLAST: Yank_9, function unknown E-value: 9e-63 LO: No
Future Work: Use the techniques and programs that we have become more familiar with to continue our progress with annotating genes.