Nice work! Sounds like you got one of the really aggressive and lycogenic ones! Exciting stuff
Author Archives: jeremysieker
Comment on The Pipette by jeremysieker
Setback
Lesson Learned
Comment on First impressions are important but every phage deserves a second chance by jeremysieker
Comment on Mine by jeremysieker
This literally makes you my new hero. That is absolutely fantastic. Be honest, how much time did you spend on that?
Final Purifications and Flooding
Yesterday, I got back my plates after two days of incubation and my third round of purifications all came back very positive. Although the colonies produced from my phage are very opaque (honestly, just weak), they are consistent and I believe that I have isolated a single strain of the bacteriophage. Therefore, I started the next step of the process, yesterday, which involves a process called “flooding” where you fill the agar plate with phage buffer and wait for a few days, then use that phage buffer (with phages in it) to do another round of plating in plaque assays. I am going in tomorrow to do that plating so that I do not lose an entire week if my phage isn’t strong enough to survive the flooding. I’m really excited to be moving forward like this.
Update
I had to redo my second round of purification because the second round came back without any presenting plaque colonies. However, it was redone and there were many plaque/phage colonies this week and I prepared my third round of purifications today and they are incubating now until Wednesday’s lab. About half the phage group has found their phage, so far. In all honesty, it seems that it was a “luck of the draw” sort of deal in terms of who cultivated a phage. However, new soil samples are being made and the search continues both with arthrobacter sp and with micobacter sm hosts.
Purification Round 2
After the first round of purification, I once again found a phage. This means that I have truly begun the process of isolating a single phage. During lab on the 30th, I did another 6 plaque assays for my second round of purification. It is a little bit funny, because at first glance my plates appear to be absolutely empty, but when you look at them with light on them at an angle you find that they are absolutely webbed with Phage Concentrations: they simply aren’t as aggressive or pervasive as many of the phages from the past. I don’t care that my phage is relatively weak, I am happy to have found one.
Results of first round of purification
Great results! On friday, after class, a few of us went into Dr. Gibbon’s lab to go check on our phage plates and the first round of purification went great! Some students had very invasive phages, but the ones that I have are much less aggressive and much more difficult to see. Despite these differences, I am still very excited to have truly begun the process of isolating a phage.
Success at last!
Today in lab, we looked at our samples and although there was a widespread problem of contamination, about 9 or 10 of us were able to form/find phage and plaque colonies! The old host was unsuccessful (Mycobacterium Smegmatus) this time, but for some reason (likely a better round of bacteria) a few of us (myself included) were able to cultivate some phage! This is really exciting because I was starting to question what we were going to do, since we had already attempted twice to no concrete avail. However, now, we are able to begin the isolation and purification process of the phages.
New Procedure
Yesterday (18 September) all of us phage-ers used a slightly different procedure. We prepared the agar plate as usual to go along with our final samples, but we also prepared another plate with 7H9+1mM CaCl_2 agar and the host bacterium that has been used in years past: Mycobacterium Smegmatus (Smeg). We spotted that new type of plate with 0, -1, -2, -3, and -4 concentrations of each of our three phage solution samples. Today (off-day for lab) I went in to go check and see if one day was possible a better time than the five day growth time that we usually allow. With the Arthrobacter host, the results are unclear. Most of the class’s plates seemed very clean; however, my plate and Walker’s plate both showed some possible phage action: his much more so than mine, mine is likely just contamination. But when I looked in the incubator at the plates for the Smeg host, many plates showed very positive signs of phage presence: a welcome sign! I am hopeful for Arthrobacter Phage activity in at least Walker’s plate and if that fails, for some progression of research with the Smeg host.
Contamination Plate
Take 3
Today in the lab, after having been given an extra five days to culture and grow, there has still been no phage to cultivate: our second try did not go as we hoped. Also, that streak test that had the interesting anti-bacterial property to it turned out to be relatively insignificant to the class’s goal, so I threw it out along with the rest of the samples: it was a cool, unique finding though! So, now, we are going for it a third time. I’m starting to wonder what about our procedure is going wrong. I have the terrifying suspicion that it is something simple that we are overlooking. Maybe we shouldn’t obey that “hypothesis” about how CaCl_2 increases phage infect-ability as if it were a law. Whatever the issue may be, others are sharing the issue: no Texas schools have cultivated any phage.
I am learning to redefine what failure is and what the real goal of this project is. I need to remember that I am doing this to research and learn, not just to find phages. Whatever it is, I am loving this learning process. However, we will need to start progressing sometime soon or we will have to revert to using the host bacterium of years past: I hope that doesn’t have to be the case
Interesting Results
The first attempt at the experiment was relatively unsuccessful: all of us learned a great deal about laboratory procedure (maybe it was better to make the rookie mistakes on a batch to be thrown out, anyways!), but none of us were able to successfully plant our phage. We tried it all again, though! A few people did find some sort of result, but those were mostly due to contamination.
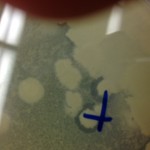
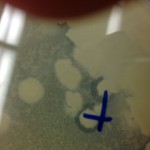
I had an interesting sort of contamination, though. When I was creating my agar plate, using the streak-test technique, I accidentally used an arthrobacer-sp tube that Carter Lantz was using to set up his Plaque-Assay experiment. Due to my mistake, the plate I made had double the bacteria and two different sets of phage concentrate in it. I though I had ruined the experiment and tried to throw it out. However, Dr. Gibbon and Dr. Adair caught me before I threw it out and encouraged me to incubate the sample and let it grow. I figured “Why not?” and I am very happy they stopped me from throwing it out. Whatever was going on with that bacteria and the phage had some interesting effects. After it grew for two days (09 September-11 September), I checked it and the bacteria had grown like crazy and around some of the major colonies of bacteria, there were areas that had become absolutely clear of everything: bacteria, plaque, agar, everything, all gone. This means that whatever happened to grow in that sample had some sort of antibiotic property to it.
For the agar to have disappeared, something must have consumed it: implying that there was something that grew there. However, looking at it on Wednesday, the whole area was clear: implying that something else killed it or it moved, something must have happened. I went back in today to go check on it (two more days later) and it appeared as if the colonies were starting to regrow over the areas that had been previously cleared.
I have no idea what is going on here, but that is the beauty of research! It may not be 100% related to the SEA-Phages project, but I hope to keep looking into whatever happened here. Maybe another research project someday!
I’ll upload the pictures in just a few minutes.