Lab 14: Poster Presentation and Abstract Submission
Lab 14: Poster Presentation and Abstract Submission
The purpose and objective:
The objective of this lab is to complete the metadata form, final draft abstract and scientific poster.
Date: 04/26/2019
Procedures:
- Fill in the form of soil metadata through Excel;
- Discuss and write the abstract, and edit the scientific poster;
- Upload file on Canvas and box.
Data:
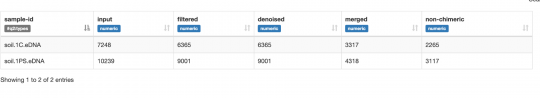
- Metadata
| Group Members | Section | Group | Soil ID | GPS location | Tree Species |
| Michelle Nguyen, Lauren Langston, Zihan Yuan | 22 | 6 | LNY22_6Sp19 | -97.11,31.55 | Ilex Vomitoria |
| BHD(cm) | pH | Soil Texture | Extraction Method | DNA Concentration/µl | Volume µl | PCR | Soil Label On Bag |
| 38.1 | 6.5 | Sandy Clay Loam | Silica Bead | 1.46 | 10 | + | LNY22_6Sp19 |
- Abstract
Isolation and Purification of Ciliate DNA from Ilex Vomitoria
Nearly 85% of ciliate diversity has not been accounted for, yet ciliates play a crucial role in soil ecosystems, ensuring the cycling of materials and energy flow of other microorganisms and serving as predators regulating the environment. This study was conducted to develop a well-defined methodology that could be used to explore soil ciliates on a molecular level via sequencing. Soil samples were taken from an Ilex vomitoria and DNA was extracted using the silica bead extraction method. Metadata analysis of the soil showed that the soil type was sandy clay loam and had a pH level of 6.5. Gel electrophoresis showed that crude DNA was extracted from the soil samples and PCR was able to amplify an 18S V4 region of the DNA. From the Nanodrop analysis, performed after the extraction of a crude DNA sample and DNA purification, the extracted DNA had a purity of 1.46 (A260/A280), which is the expected score of a pure sample. The utilization of this less costly and more efficient protocol can yield DNA samples of a good purity that can be used for future sequencing and taxonomic identification of organisms within a collected sample.
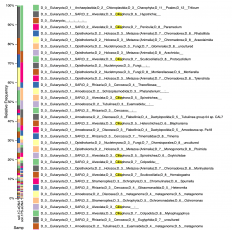
- Poster

Conclusion and The future goal:
In this lab, based on discussion, suggestions and comments, we edited and finished our final abstract and poster successful. In the future, we will do a presentation of our scientific poster next week and I am looking forward it!