2/21/19
Objective/Goals: The objective of this lab was to effectively set up the gel electrophoresis box with our samples. The goal was to determine if there is any DNA in our samples, and if so, how much.
Materials: Gel box, DNA sample, 1 x TAE buffer, micropipettor, ladder substance, tube
Procedure:
Gel Electrophoresis
1. Make sure Gel is placed in buffer chamber and covered with the 1 x TAE buffer
2. For each sample, mix 9 μl of the DNA sample with 1 μl of the 10x loading buffer
3. Mix tube well and repeatedly tap on table
4. Load 10 μl of sample in lane 7
5. Load 5 μl of “31” 6.2 ng/μl in lane 6
6. Load 5 μl of “62.5” 12.5 ng/μl in lane 8
7. Run gel for 20-30 minutes at 100v
Gel Imagery
- Take gel out of electrode box
- Transfer gel to upstair lab
- Place gel block onto tray and insert into the gel imager
- Take pictures of the gel block with the visible DNA bands
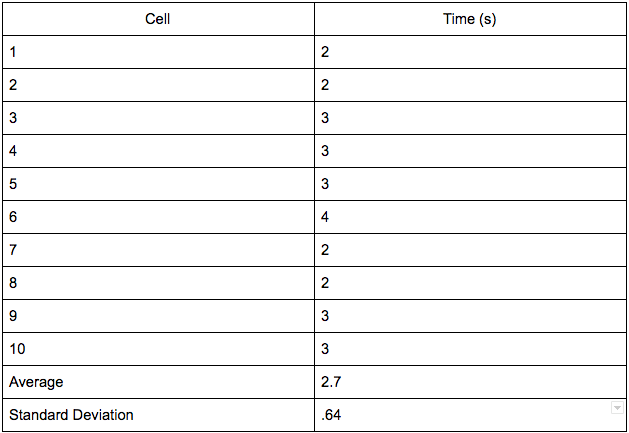
Nanodrop
- Clean surface of the nanodrop machine with DI water
- Add one drop of DNA sample to machine
- Close lid
- Record data from screen
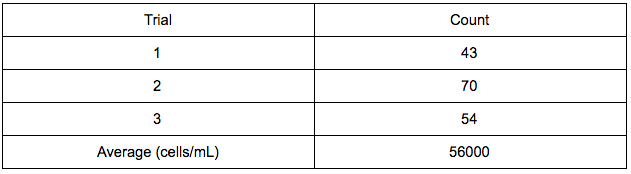
Data:
Nanodrop-
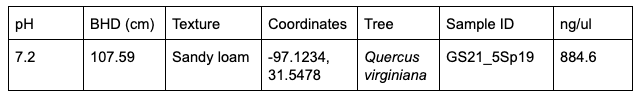
884.6 ng/μl
1.44 A260/A280
0.41 A260/A230
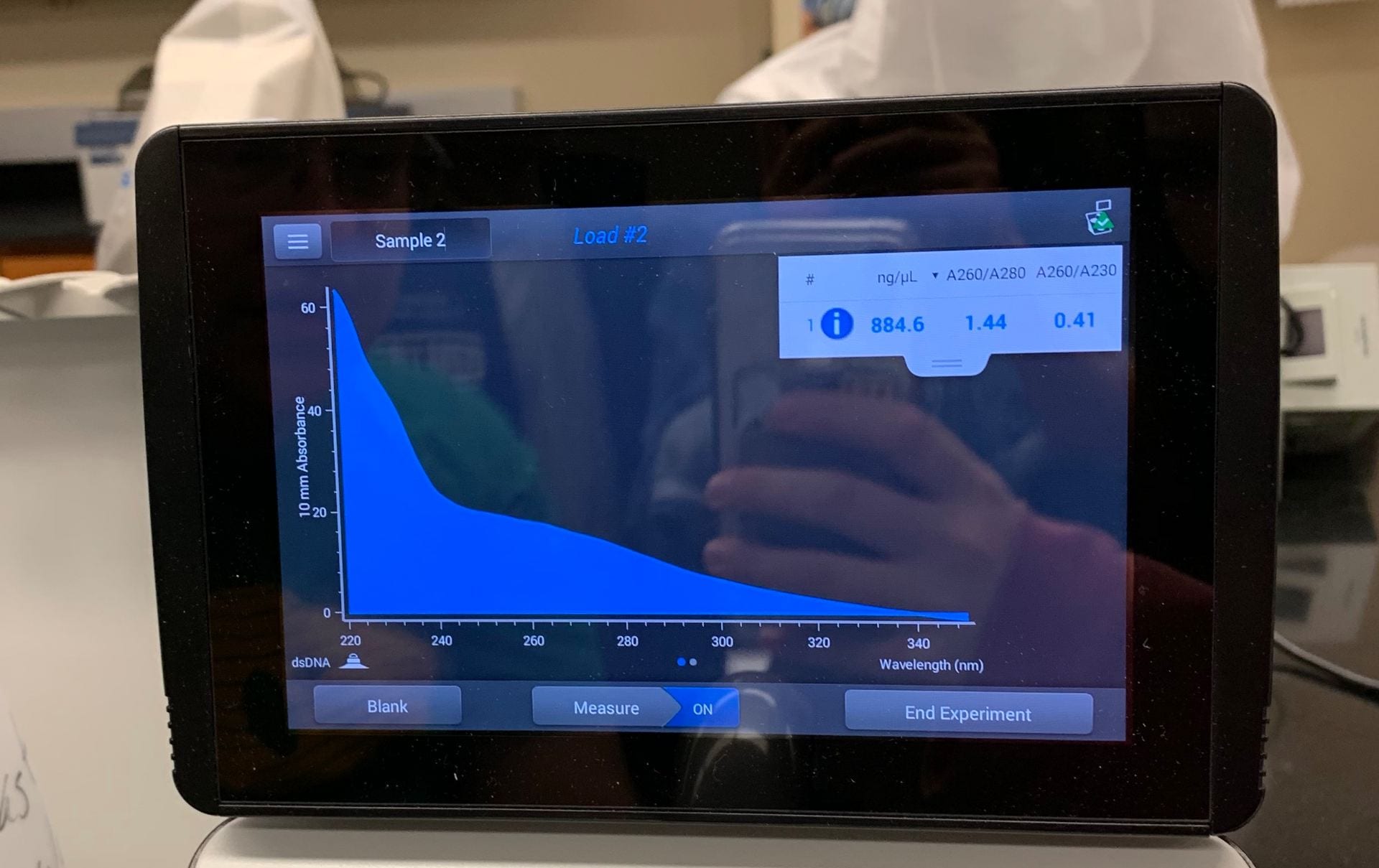
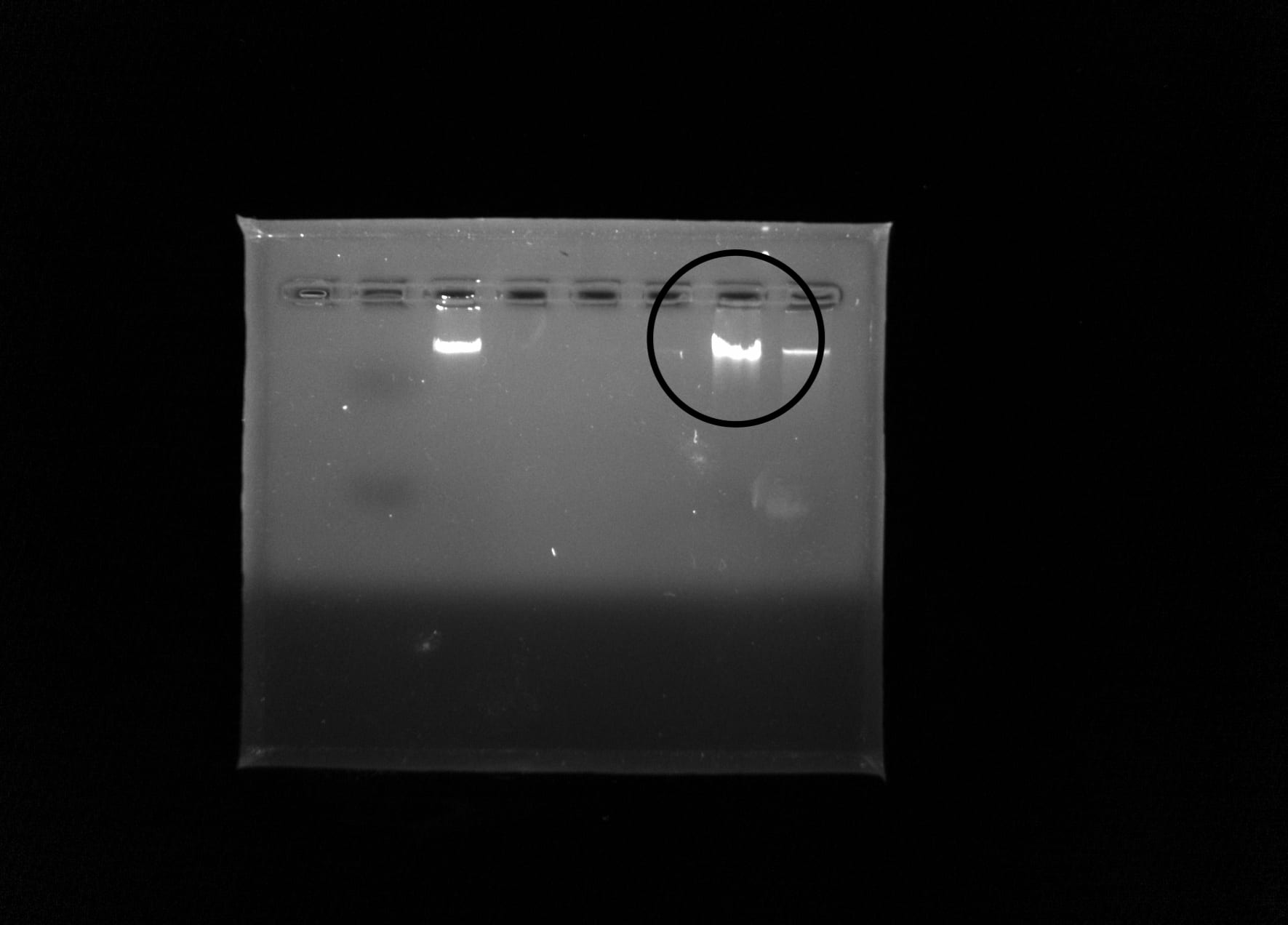
Observations: A bright band of DNA showed up in my groups lane in the gel. However, the nanodrop analysis showed that our DNA sample was not very pure.
Storage: The gels were disposed and the boxes were emptied of water. All of the DNA sample tubes were placed back in the cooler.
Future Goals/Conclusions: Overall, I think it was successful that my group had some DNA, even though it was not as pure as we had hoped for; I did not expect the sample to be very pure because it was light brown in color. In the future, I would like to be able to secure a strong DNA sample and make sure it is very pure.