Shivam Patel 4/25
Purpose: The purpose of the lab was to create abstracts for our poster and finish our posters, as well as label and sort our soil.
Procedure:
1. Label soil bag with your sample ID.
2. Fill in the excell spreadsheet with information of your sample.
2. Finalize abstract and poster.
Abstract:
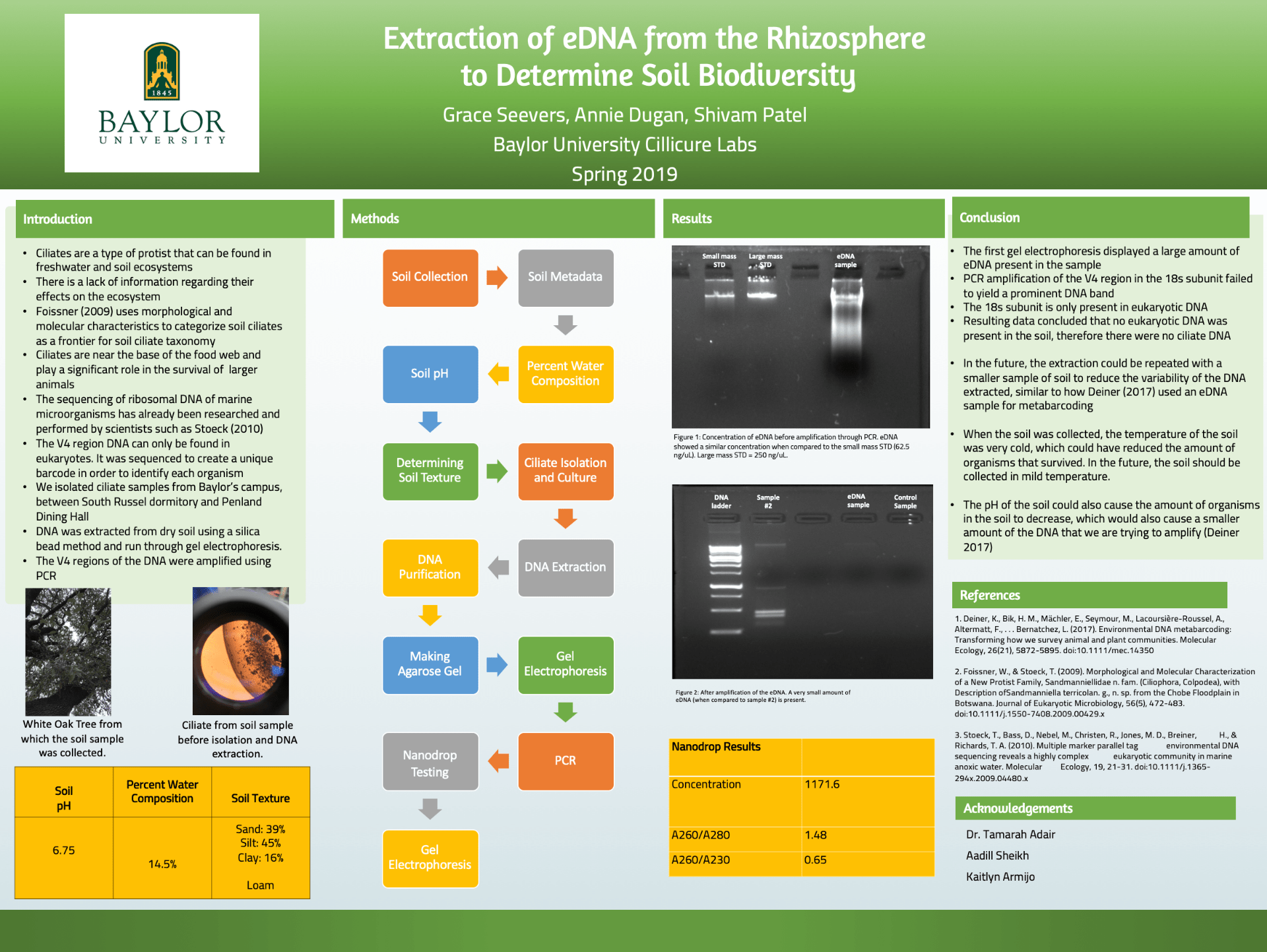
Ciliates are unicellular eukaryotic organisms that reside in freshwater or soil ecosystems. Ciliates are an important factor within the ecosystem because they consume different bacteria and protozoa, and act as a source of nutrition for organisms at higher trophic levels. The enormous discrepancy between the amount of research conducted on ciliates living in marine biomes versus ones living in the soil is due to a general lack of interest and/or knowledge of their importance. This experiment was conducted over the course of fourteen weeks in an effort to fill the gap between the amount of information known about marine and soil biodiversity by determining the overall composition of ciliates living in the rhizosphere of different trees across Baylor University’s campus. The V4 region of the 18S rRNA was observed because it is conserved within all eukaryotes and is unique to each specific organism. Soil samples were collected from the rhizosphere, where the highest concentration of ciliates are presumed to inhabit because of its abundance of nutrients and gas availability. After collection, the soil was searched for the presence of ciliates. Once the ciliates were isolated, their DNA was extracted and purified. The pure DNA was amplified through a polymerase chain reaction and ran through gel electrophoresis to determine whether or not eukaryotic DNA was present. Although our sample contained a very minute amount of DNA, if substantial DNA had been found, it would be sequenced using an online bioinformatics platform- QIIME2, to determine the specific organisms in our sample.
Conclusion: With our poster and abstract finished, now we just need to prepare to present at the symposium.

