Lab 8: PCR Results and Scientific Poster Design
Objective:
The main goal of this lab was to run our samples through gel electrophoresis in order to determine whether or not our samples qualified to be sent off to be sequenced. Furthermore, we took our midterm which allowed us to review everything we have covered so far in lab and make sure we remember it well. Lastly, we began drafting our scientific poster that we will present later in the semester.
Procedure:
- The 1.5% gel was previously prepared and already set up for us to begin pipetting
- Pipette 5 µl of 1 kb ladder into well #1 of the gel
- Pipette 10µl of your DNA sample into well #2
- Pipette 10µl of your control sample into well #3
- Then the other group’s control and DNA sample were pipetted into the next two wells
- Run the gel at 100V for 30 minutes
- Use the imaging software to view the results of gel
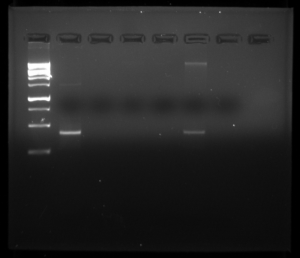
Data:
The first image shows our results from the imaging software. The second image shows our wells filled with our DNA and control groups.
Conclusion/Future Goals:
The results show that our DNA was pure enough to show a high concentration on the imaging. One of the other two groups results also showed a high concentration of DNA. In the future, our DNA could be sent off to be sequenced. Furthermore, we are going to continue working on our scientific poster in order to have it ready to present when the time comes.