Lab 14: Poster Presentation and Abstract submission 04/25/19
Objective:
The objective of this lab was to organize the entire classes sample data through bringing together multiple aspects of the soil in a single excell spreadsheet. It was also to revise our poster, poster title, and abstract.
Purpose:
The purpose of this lab was to allow for a clearer picture of the overall results to be achieved through assimilating information together across multiple samples. It was also to spend additoinal time dedicated towards creating a better poster and abstract and to have access to the instructors for questions.
Procedure:
- Vote on a logo.
- Enter soil metadata results into the excel spreadsheet for your class.
- Communicate your sample ID to the instructor after identifying the remainder of your group’s extracted DNA sample.
- Revise your poster, poster title, and abstract while using the instructors as needed.
Data/Observations:
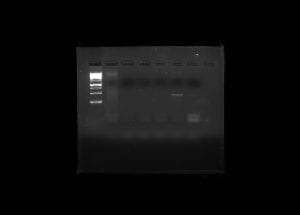
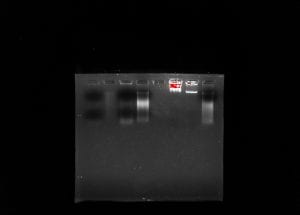
Due to our PCR results being negative, our metadata was taken off the spreadsheet and considered to contain no importance.
-Title-
Soil eDNA Analysis Targeting the V4 Region Using the 18S Primer
-Abstract-
Soil ciliates are a diverse group of eukaryotes that play an important role in the environment but are understudied due to a lack of standard methods used to extract them from soil. The purpose of this experiment was to determine ciliate biodiversity in soil samples taken from the rhizosphere of Baylor trees. The protocols used were ones that have yet to be perfected in hopes of establishing them as universal method such as silica bead extraction of DNA and the chelex extraction method. DNA was extracted and purified from the soil collected using the ‘silica bead method’. The DNA concentration was obtained using a nanodrop and gel electrophoresis by running against a mass standard, and used PCR to amplify the 18s V4 region. The Polymerase Chain Reaction yielded a negative result, indicated by a ‘smear’ of DNA rather than a band. The nanodrop showed the concentration of DNA to be 262.1 ng/µl with the A260/280 to be 1.5. Despite negative results, the experiment showed the validity of results that can be found through silica bead and chelex DNA extraction; however, the lack of positive results was most likely due to an improper dilution or a degradation of DNA. Additionally, further research can be done using bioinformatics to visualize the ciliate diversity.
-Poster-
Conclusion:
In conclusion, this lab proved to be important in organizing the data across the different samples and to see which showed positive PCR results. This occurred for 10 samples and it will soon be decided which ones will be chosen go into further detail with. It also served to allow the quality of our material for the CURES symposium to be improved.
Storage:
There was no storage needed for this lab. Extracted DNA samples were relabelled and our desks were cleared to ensure it was left as it was found.
Future Goals:
In the future, I hope to conduct bioinformatics on the samples that showed positive results after PCR. After this, I hope to discover the biodiversity with the rhizophere of Baylor’s trees around campus and observe the types of ciliates within it.