Garrett Darden
Biology 1106-Section 25
Dr. Adair
4/26/2019
CILI-CURE Lab #14: Poster Presentation and Abstract Submission
Objective: The objective of lab this week was to begin preparing for the CURE Symposium which will take place next Friday. We did that this week by once again correcting our posters, writing our abstracts, and organizing our soil metadata into a spreadsheet.
Purpose: The purpose of this lab was to set everything logistically in place for CURE Symposium next week, like prepping our posters for printing and creating abstracts that will be printed into the pamphlet for the event. The second purpose was for us to once again look at our older data and to create an abstract as well as further improve our posters
Materials:
- Computer with Internet Connection
- Access to Prior Notebooks
- Soil Bag
- Flask with Post-PCR DNA Sample
- Access to Copy of Poster
- Access to Writing Program like Word or Google Docs for Writing the Abstract
Procedure:
- Open up your computer and follow the metadata link to the metadata spreadsheet
- Use your group members, soil bags, post-PCR DNA sample flask, and older notebooks from online or your personal notebook in order to fill out all of the blanks on the spreadsheet.
- Look up older data like GPS coordinates, BHD, the nanodrop data, tree species, soil and pH assays, and the soil bag ID and the estimated volume of post-PCR DNA sample flask
- Once all of the data is tranferred over to the spreadsheet, return the soil bags and DNA sample flask to their respective containers, and clean up the lab space and move to a computer lab for the next steps.
- Prior to the lab, draft abstracts should have been made by each individual member of the group, so now pull up each draft and work together in order to combine the three into one abstract that will represent the whole group and aim to have the draft be at max 250 words.
- Now ask your TA’s and instructor what suggestions and critiques they have of your poster and be sure to write them down so that they can be corrected later.
- Whether or not the title is changed, put the final title of the poster with the final abstract and upload the paper to the designated BOX folder as well as too Canvas under the QTM 14 assignment.
- Once that is finished then work as a group in order to address the suggestions and critiques given by your TA’s and instructor in order to improve and fix the poster.
- After all comments and suggestions have been addressed and the poster is finished, upload the file to the BOX folder so that it can be printed for next week.
- Shut down the computer and complete the paper QTM and turn it in to your TA’s prior to leaving the lab and clean up your lab space prior to leaving.
Results/Data:
Soil eDNA Metadata Spreadsheet:
Group Members: Garrett Darden, Caleb Touchstone, and Chris Amezcua
Section: 25
Group: 2
Soil ID: ADT25_2Sp19
GPS Location: (-97.1136, 31.5445)
Tree Species: Quercus Virginiana
BHD (cm): 44.03
pH: 4.1
Soil Texture: Clay Loam
Extraction Method: Silica Bead
DNA Concentration ng/microliter: 250.2
Estimated Volume: 50
PCR: +
Soil Label on bag: CT25Sp19
Final Title and Abstract:
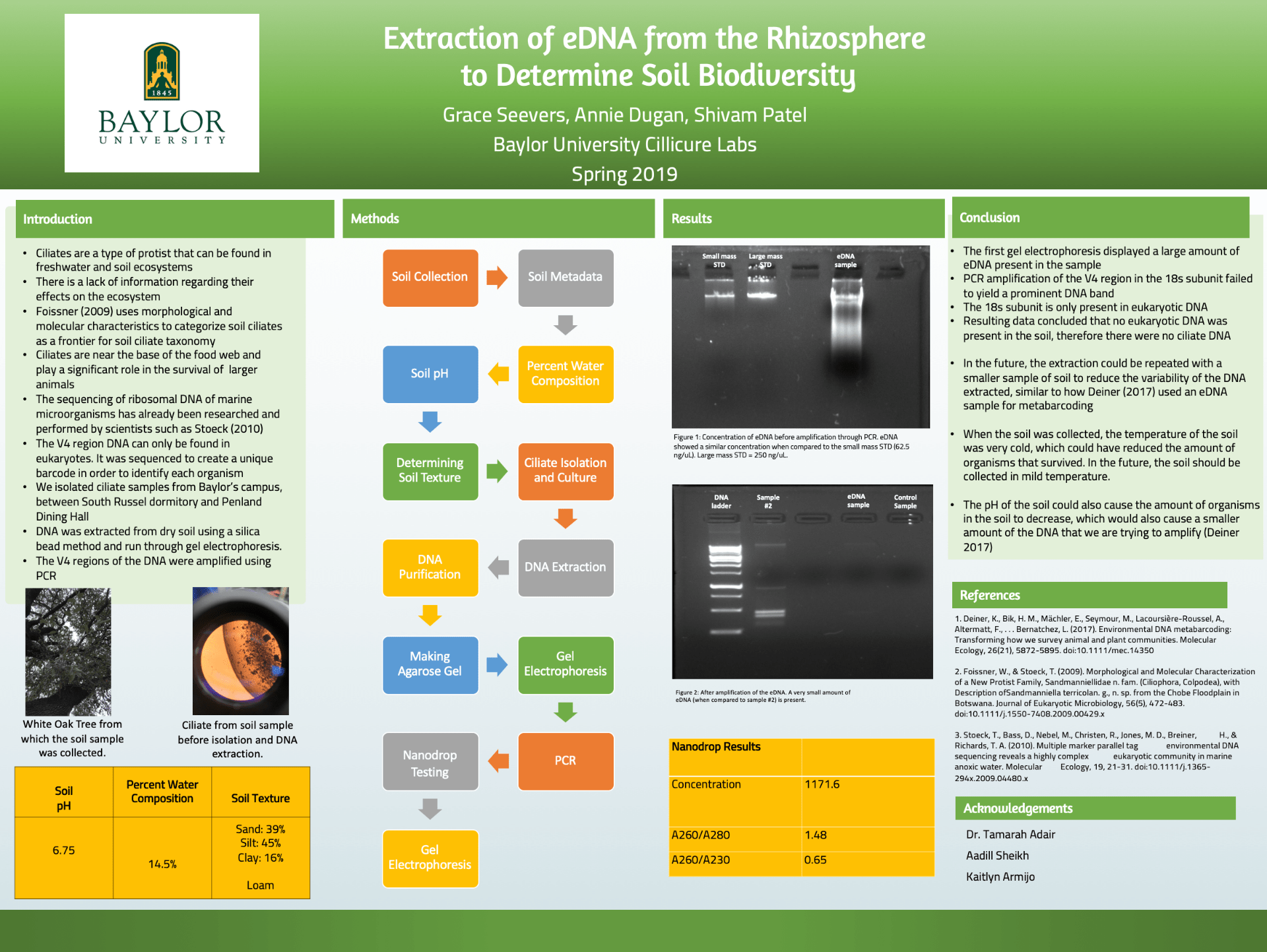
Title: Analysis of eDNA from a Soil Sample in the Search for Soil Ciliate DNA
Abstract: Soil ciliates are crucial organisms in the rhizosphere as they perform many important roles contributing to soil health. Currently, research concerning topics related to soil ciliate diversity is limited. Conventionally, ciliates have been identified and categorized through morphological characteristics and behaviors due to a lack of a standard genomic method for easy identification. This study aims to explore a standard method for ciliate identification through metabarcoding. For this, soil was collected and environmental DNA was extracted using the Silica Bead grinding method followed by Column Purification. Polymerase Chain Reaction was used to amplify the V4 region of the 18s rRNA subunit. Gel electrophoresis results showed that PCR amplified the eDNA. The image taken of the gel showed a concentrated band of DNA from the eDNA sample. Nano-drop analysis of the eDNA showed a 250.2 ng/ml of the DNA while it had a purification of 1.46 A260/280. The eDNA extraction method conducted appears to be the best option for extracting DNA due to the amount of DNA replicated. Metabarcoding of the V4 region of the 18s rRNA subunit was conducted using the eDNA to identify what kind of ciliates were extracted from the environmental sample and sequence. Bioinformatics (Cyverse/QIIME2) will be used to analyze how successful the V4 region can be in the identification of ciliates.
Copy of the Poster: 
Conclusion: In conclusion, I believe that this week was insanely productive because we were able to put the finishing touches on the poster as well as create our abstracts. I also began to feel more and more comfortable talking about the content of my poster to others when talking through it for suggestions and critiques
Future Goal: In future labs I want to continue improving my poster creation and abstract writing skills so that I can continue to deliver research quality work for such events as CURE Symposium and and Scholar’s week. I also want to be able to continue running metabarcoding so that we can continue to look at the diversity of the eukaryotes in the soil sample to a deeper degree, more so than we have in the past few weeks while working with Qiime2 and CyVerse.