Annotation Day 4: Nubia Genes 53, 54, and 55 [02/15/17 Cori Hughes]
Journal entry by Cori Hughes on the 15th of February, 2017
Goals: annotate at least two of Nubia’s genes (starting at gene 53 and moving downstream)
Tools: HHPred, Starterator, DNAMaster, GeneMark, PhagesDB BLAST, NCBI BLAST
Results for Gene 53:
Start: 38240bp Stop: 38461bp FWD GAP: bp SD Final Value: SD Score: -6.414 (4th best score) Best score not chosen. Best score (-3.714 is much shorter in length and does not cover all the coding potential) Z-Value: 1.433 CP: The gene is covered SCS: Agrees with Glimmer, Agrees with GeneMark NCBI BLAST: hypothetical protein RAP15_52 [Arthrobacter phage RAP15] E-Value: 1e-44 CDD: No good hit PhagesDB BLAST: GreenHearts_Draft_55, function unknown E-Value: 4e-36 HHPred: No good hit LO: Yes ST: Agrees with Starterator F: NKF FS: NCBI, PhagesDB

GeneMark covers 38240bp-38461bp (the middle frame).

HHPred results show no good E-value.

NCBI BLAST show an alignment with hypothetical protein RAP15_52. The query cover is 98% and the E-value is 1e-44.

This is the query:subject from the NCBI BLAST alignment with hypothetical protein RAP15_52.

PhagesDB BLAST shows a significant alignment with Greenhearts_Draft_55 with an E-value of 4e-36.
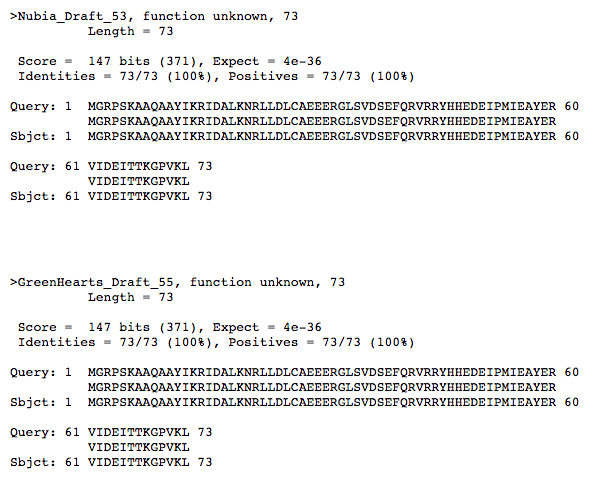
This is the query:subject of both Nubia_53 and its closest alignment on PhagesDB BLAST, GreenHearts_Draft_55.

Nubia gene 53 matches the suggested start location on Starterator.

These are the start locations of similarly matched genomes to Nubia’s gene 53.
Results for Gene 54: Not completed. Due to need for revision and review, gene 54 will be looked at as soon as possible.
Results for Gene 55:
Start: 39753bp Stop: 39439bp BKWD GAP: 63bp Gap SD Final Value: SD Score: -5.525 (3rd best score) Best score not chosen. Best score (-1.984 is too short and did not cover all coding potential) Z-Value: 1.543 CP: The gene is covered SCS: Disagrees with Glimmer, Agrees with GeneMark We agreed with GeneMark because it is the longest ORF NCBI BLAST: hypothetical protein SEA_OXYNFRIUS_53 [Arthrobacter phage Oxynfrius] E-Value: 2e-55 CDD: No good hit PhagesDB BLAST: Oxynfrius_53, function unknown E-Value: 6e-45 HHPred: No good hit LO: Yes ST: The start location was changed to 39753. The ORF length is the longest but does not have the best SD score or the best Z value. F: NKF FS: NCBI, PhagesDB

HHPred did not show any good hit for Nubia gene 55.

NCBI BLAST shows a hit for hypothetical protein SEA_OCYNFRIUS_53 with an E-value of 2e-55.

These are the query:subject on NCBI BLAST for hit hypothetical protein SEA_OXYNFRIUS_53 and the next-best hit, hypothetical protein JOANN_54.

PhagesDB shows a hit for Oxynfrius_53 with an E-value of 6e-45.

PhagesDB BLAST query:subject for Oxynfrius_53 and the gene being blasted, Nubia_Draft_55.

The suggested start for Nubia_Draft_55 (39696) on Starterator was not chosen. Instead, 39753 was chosen to have the longest ORF length.

Conclusions: Gene 53 and Gene 55 of Nubia were completed. Gene 55 has a different start location than what was suggested on Starterator in order to have the longest ORF. We (Roshni and I) have started annotating backward genes.
Next Step: Annotation Day 5: Nubia Genes 54, 56, 57, and 58 [02/20/17 Cori Hughes]